Abstract
Since 2012, the clinical cases of inclusion body hepatitis showed an increasing trend in China, causing considerable economic losses to the poultry industry. In this study, a fowl adenovirus strain CH/GDLZ/201801 was isolated from a chicken flock experiencing inclusion body hepatitis and analyzed by complete genome sequencing. The pathogenicity of the new virus strain was examined by experimental infection of specific pathogen free chickens. The isolate was identified by immunofluorescence and the virions presented typical icosahedral particles under transmission electron microscopy. The full genome of the isolate was 44,329 nucleotides in length with 58% G+C content. Phylogenetic analysis, based on the whole genome, revealed that the new isolate was closest to serotype 8a from the species Fowl aviadenovirus E (FAdV-E). Recombination analysis and phylogenetic analysis showed that the new isolate is a recombinant strain between FAdV-8a and FAdV-8b. In infection experiments, three infected chickens showed clinical signs and one chicken died on day 7 post infection, corresponding to 5% mortality. Macroscopic and microscopic lesions in the liver were observed, and viral antigen could be detected in the livers by immunohistochemical staining and TEM. Taken together, our study describes the genomic characteristics and pathogenicity of a FAdV-8a strain in China. It would lay a solid foundation for further study of the pathogenic mechanism and vaccine development of the virus.
Similar content being viewed by others
Introduction
Fowl adenoviruses (FAdVs) are non-enveloped double-stranded DNA viruses with a genome of 43–45 kb in size, belonging to the genus Aviadenovirus in the family of Adenoviridae (Harrach et al. 2011). The viruses are clustered into 5 species (FAdV-A to FAdV-E) based on restriction enzyme digestion patterns (Harrach et al. 2011), and they are further classified into 12 serotypes (FAdV-1 to 8a and 8b to 11) by cross-neutralization tests (Hess 2000). In addition, FAdV-1 belongs to the species FAdV-A, FAdV-5 belongs to the species FAdV-B, FAdV-4 and 10 belong to the species FAdV-C, FAdV-2, 3, 9 and 11 belong to the species FAdV-D, FAdV-6, 7, 8a and 8b belong to the species FAdV-E (Harrach et al. 2011). The genome of FAdV encodes a number of non-structural proteins and three major capsid proteins: hexon, penton base and fiber. The hexon protein, as a major surface-exposed capsid structure, harbors group-specific, type-specific and subtype-specific antigenic determinants (Toogood et al. 1989; Russell 2009). The penton base forms the vertices of adenovirus capsid in association with some other proteins, and plays a major role during penetration and entry of the virus into the cell (Fender et al. 2005). The fiber protein is responsible for the primary cell attachment and might also be involved in pathogenicity (Russell 2009; Nicklin et al. 2005; Pallister et al. 1996).
FAdVs are transmitted both vertically and horizontally, and widely distributed in poultry industry throughout the world (Schachner et al. 2018; Grafl et al. 2012; Grgic et al. 2006). Most of FAdVs are opportunistic pathogens and their infections are subclinical (Choi et al. 2012). However, some of FAdVs are associated with specific avian diseases including inclusion body hepatitis (IBH), hepatitis-hydropericardium syndrome (HHS) and gizzard erosion and ulceration (GEU), causing significant economic losses to poultry industry (Mase et al. 2012; Domanska-Blicharz et al. 2011; Grafl et al. 2012; Lim et al. 2011). All 12 serotypes of FAdVs have been associated with outbreaks of IBH (Lim et al. 2011; Winterfield et al. 1973). The primary lesions of IBH include congested and enlarged liver with hepatic necrosis, and eosinophilic or basophilic intranuclear inclusion bodies in hepatocytes (Mase et al. 2012; Ojkic et al. 2008). In addition, pancreatitis has been observed in some cases of IBH in chickens infected experimentally with FAdV-1 and FAdV-8 (Gallina et al. 1973; Grimes et al. 1977; Grimes and King 1977).
In this study, a FAdV strain was isolated from Guangdong Province of China. The whole genome sequence of the strain was characterized, and its pathogenicity in 4-week-old SPF chickens was investigated. These results suggest that the isolate of FAdV CH/GDLZ/201801 is closely related to FAdV-8a and caused 5% mortality in SPF chickens.
Materials and Methods
Sample Collection and FAdV Detection
A suspected clinical case of inclusion body hepatitis occurred in a 4-week-old broiler chicken flock in 2018 in Lezhu Country, Guangdong Province, China. Liver tissue from dead chicken was collected and homogenized in phosphate buffered saline (PBS) to obtain a 10% tissue suspension. The suspension was centrifuged at 5000 ×g for 10 min after three freeze–thaw cycles, and the supernatant was used for DNA extraction by using the TIANamp Genomic DNA Kit (TIANGEN, Beijing) according to manufacturer’s instructions. The presence of FAdV-8a in the liver sample was confirmed with PCR and sequencing analysis by Beijing Genomics Institute (Guangzhou, China). PCR was performed using the primer pairs FAdV-F: 5′-TACTGCCGTTTCCACATT-3′ and FAdV-R: 5′-CGGTGTTCACGATAGCC-3′ to amplify an approximately 1431 bp product from the hexon gene, as previously described by Ruan et al. (2017). Other viruses including Marek’s disease virus (MDV), Newcastle disease virus (NDV), avian influenza virus (AIV) and infectious bursal disease virus (IBDV) were examined by PCR or RT-PCR using primers and methods as previously described (Liu et al. 2013, 2014; Lu et al. 2015).
Virus Isolation and Purification
The FAdV-8a positive supernatant was propagated on Leghorn male hepatoma (LMH) cells (ATCC #CRL-2117) for virus isolation (Kawaguchi et al. 1987). Briefly, LMH cells were seeded into 6-well plates, and maintained in DMEM F12 medium (Hyclone, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and 1% penicillin–streptomycin (Gibco, USA) at 37 °C with 5% CO2. The FAdV-8a positive supernatant of liver lysate was filtered through a 0.22-µm-pore-size syringe filter (Millipore, USA) and used as an inoculum for virus isolation. Filtered inoculum was diluted tenfold in DMEM F12 medium before inoculation onto the LMH cell monolayer. After 1.5 h of absorption at 37 °C, the supernatant was removed, and 2 mL DMEM F12 medium, supplemented with 2% FBS was added to the monolayer in each well. The cells were cultured continuously at 37 °C in 5% CO2 atmosphere. When extensive cytopathic effects (CPEs) were observed, the supernatant and cells were harvested by three freeze–thaw cycles, followed by centrifugation at 1500 ×g at room temperature for 10 min. And the supernatant was used as seed stocks for the plaque purification.
The virus was plaque-purified three times as described previously (Alexander et al. 1998; Marek et al. 2010). Briefly, the supernatant was serially diluted and used to inoculate LMH cells in the DMEM-F12 medium for 1 h at 37 °C in 5% CO2, and then the culture supernatant was discarded. The medium was then replaced with a 1:1 mixture of 2% Agarose LM GQT (TaKaRa, Dalian) and DMEM F12 medium. After 3–5 days of incubation at 37 °C in 5% CO2, the isolated plaques were picked and the virus was propagated in 6-well plates until CPE was evident. The selected plaque of the virus was used for viral propagation. Subsequently, LMH cells were cultured in T175 flasks, and washed three times with sterile PBS. One mL of FAdV-8a together with 50 mL DMEM-F12 medium was added into the flask, and then was cultured at 37 °C in 5% CO2 to observe CPE. When CPE was evident, the plates were frozen at − 80 °C and thawed twice. The supernatant were harvested to determine viral titers.
TCID50 Assay
The TCID50 assay was performed with a few modifications as previously described (Steer et al. 2015). LMH cells were seeded in 96-well plates at a density of 105 cells per well and cultured overnight. Before inoculation, the monolayer was washed twice with PBS, and 0.1 mL of tenfold serial virus dilutions was added to the cells in eight replicates per dilution. Consequently, 0.1 mL DMEM F12 medium, supplemented with 2% FBS was added, and the cells were further incubated. Viral CPE was observed for 5–7 days, and virus titer was calculated as the 50% tissue culture infective dose (TCID50) using the Reed-Muench method (Reed and Muench 1938).
Immunofluorescence Assay (IFA)
Immunofluorescence assay was conducted to observe FAdV-8a-infected LMH cells as described previously (Wang et al. 2018). Briefly, LMH cells were seeded on 6-well plates at a density of 2 × 106 cells per well and cultured overnight, and then infected with FAdV-8a at a multiplicity of infection (MOI) of 1. At 72 h after inoculation, the cells were fixed in 4% paraformaldehyde for 15 min and then permeabilized with 0.2% Triton X-100 for 15 min at room temperature. The cells were then blocked with 1% bovine serum albumin (BSA), and incubated with FAdV-8a fiber protein specific monoclonal antibody (Wen’s Foodstuffs Group Co., Ltd, China) (1:250), followed by Cy3-labeled goat anti-mouse secondary antibody (KPL, USA) (1:500) for 1 h. Cell nuclei were stained with 4′-6′-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich), then the stained cells were photographed under a scanning confocal microscopy (Leica TCS-SP5).
High-Throughput Sequencing
The whole genome of the new isolate was sequenced by HiSeq™2000 Illumina. Briefly, genomic DNA was extracted from LMH cells using the QIAamp Mini DNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Qualified genomic DNA was fragmented with G-tubes (Covaris) and end-repaired to prepare DNA template libraries. Reads longer than 500 bp with a quality value over 0.75 were merged together into a single dataset. Next, the hierarchical genome-assembly process (HGAP) pipeline was used to correct for random errors in the long seed reads (seed length threshold 6 kb) by aligning shorter reads from the same library against them (Chin et al. 2013). The resulting corrected, preassembled reads were used for de novo assembly using Celera Assembler with an overlap-layout-consensus (OLC) strategy (Myers et al. 2000). The Quiver consensus algorithm was used to validate the quality of the assembly and determine the final genome sequence (Chin et al. 2013).
Recombination Analysis
The recombination analysis of the new isolate and its five parental strains were performed by SimPlot version 3.5.1. The nucleotide identity comparison was carried out using the Kimura (2-parameter) method with a transition-transversion ratio of 2, and the window width and step size were 200 and 20 bp, respectively.
Phylogenetic Analysis
Comparison and analysis of the genome sequences, hexon proteins, fiber proteins and ORF19 of the strain and the reference FAdV strains were conducted using EditSeq and MegAlign programs. After aligning the genome sequence of the strain with the reference FAdV sequences using the Clustal W multiple alignment algorithm, phylogenetic trees were constructed by the neighbour-joining method using MEGA version 7.0 based on the complete genomic nucleotide sequences, amino acid sequences of hexon gene, fiber gene and ORF19 from the new isolate together with other different reference FAdVs, respectively. The bootstrap values were determined from 1000 replicates of the original data.
Experimental Infection with the FAdV-8a Strain in SPF Chickens
Forty-six 4-week-old SPF chickens were randomly divided into two groups of 23 chickens each and housed separately in isolators. One group was inoculated intramuscularlly with 2 × 106 TCID50 per bird of the strain CH/GDLZ/201801. Another group was inoculated intramuscularlly with non-infected tissue culture supernatant and used as control. On the one hand, total twenty chickens in each group were observed daily for clinical signs of depression, dull feathers, crouching position and reluctance to move. In addition, the mortality of the twenty chickens in each group was recorded daily. Fecal swabs were collected from each chicken before (0 day post infection, dpi) and after challenge from 2 to 14 dpi every other day, and were submerged into 1 mL sterile PBS immediately after collection. On the other hand, three chickens from each group were necropsied at 4 dpi. At necropsy, the fresh samples (heart, liver, spleen, lung, kidney, bursa of fabricius, pancreas, small intestine, proventriculus and gizzard) were collected. One part of the samples were frozen and stored at −80 °C until being used in the DNA distribution analysis, whereas the other part of the samples was formaline-fixed for histopathology and immunohischemistry analysis.
Histopathology and Immunohistochemistry
At necropsy, fresh tissue samples were collected separately, and fixed in 10% formalin for 72 h at room temperature. Those fixed samples were dehydrated in graded ethanol, embedded in paraffin, cut into 4 μm sections, and mounted onto glass slides. Afterwards, the sections were deparaffinized, rehydrated, and stained with hematoxylin and eosin (HE), the slides were examined and analyzed with light microscopy.
For immunohistochemical investigations, 4-µm-thick tissue sections were placed on positively charged glass slides. The slides were air dried, deparaffinized, and then rinsed and incubated with target retrieval solution. After being blocked with 1% bovine serum albumin (BSA), the sections were incubated with FAdV-8a fiber protein specific monoclonal antibody (1:200) as the primary antibody for 12 h at 4 °C. The HRP-labeled goat anti-mouse IgG secondary antibody (Abcam, UK) was applied to the samples for 1 h at room temperature. The reaction was visualized with a 3,3′-diaminobenzidine (DAB) substrate kit (Dako, Denmark). Sections were counterstained with hematoxylin, dehydrated, and mounted in organic media. Tissues of chickens from negative control groups were used as negative control samples.
Real-Time PCR Analysis
Rectal swabs and various tissue samples were tested by a FAdV-8a hexon-gene based real-time PCR (qPCR), including viral standards with known plasmid concentration for quantification. Briefly, the homogenates from various tissues from each chicken were centrifuged at 6000 ×g for 5 min, respectively. Specific primers for the conserved regions of the FAdV-8a hexon gene (forward primer: 5′-CTACCCTAATCCCAACTC-3′; reverse primer: 5′-TGTCCATGACGTAGTAAG-3′), and probe (5′-FAM-ACTGA GAGCCGTCGTTCTTCA-BHQ-3′) were synthesized by TaKaRa (Dalian, China) as described previously (Niczyporuk and Czekaj 2018). Real-time PCR assay was performed on the Applied Biosystem 7500 Fast instrument (Life Technologies, USA). The PCR was performed in a 20 μL volume containing 10 μL of qPCR Mix, 2 μL of DNA, 0.04 μL 50 × Rox reference dye (Toyobo, Japan), 0.2 μmol/L of probe, and a 0.3 μmol/L of each hexon gene-specific primer. The thermal cycling parameters were as follows: 95 °C for 20 s; 40 cycles of 95 °C for 3 s, 60 °C for 30 s. A standard curve was generated by construction of plasmids. Briefly, the hexon gene was amplified from FAdV-8a strain, and the PCR products were cloned into the pMD19-T (TaKaRa, Dalian). The known plasmid concentration was tenfold serially diluted for generating a standard curve in each plate. The quantity of FAdV-8a viral DNA in tested samples was calculated based on the cycle threshold (Ct) values for the standard curve.
Transmission Electron Microscopic (TEM) Observation
Transmission electron microscopy (TEM) was conducted to observe virus particles as described previously (Niu et al. 2017; Zhang et al. 2016). The purified virus pellets were resuspended in PBS buffer and negatively stained with 1% phosphotungstic acid for 1 min. After blotting and drying, the grids were examined with a TEM (JEM-1400, JEOL Ltd., Japan).
For visualization of the viral particles in the liver of the experimentally infected chickens, portion of liver and pancreas was fixed in 5% glutaraldehyde fixative prepared in PBS buffer for 3 weeks. The samples were washed three times with PBS, the fixed tissue was dehydrated with an ascending sequence of ethanol (30%, 50%, 70%, 90%, 100%) and dried by critical point drying method. Ultrathin sections were prepared using ultramicrotome, collected on 200-mesh copper grid (PELCO) and stained with uranyl acetate for 15 min and lead citrate for 5 min. The ultrathin sections were screened by TEM (JEM-1400, JEOL Ltd., Japan).
Results
Virus Isolation and Identification
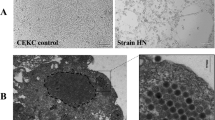
A 4-week old chicken with a clinical diagnosis of IBH was used to identify the viral pathogen causing the infection. Liver tissue sample was collected and identified by PCR. It indicated that the liver sample was FAdV-8a positive (Supplementary Figure S1). During serial passage in cell culture, the LMH cells became round and clustered like grapes at 3 dpi as compared with the control (Fig. 1A, 1B). To confirm FAdV-8a replication in LMH cells, viral DNA was extracted from the inoculated cells at 3 dpi and tested by specific real-time PCR. This cell culture-passaged sample was positive for FAdV-8a. Plaque-purified FAdV-8a in LMH cells was further confirmed by IFA with FAdV-8a fiber protein specific monoclonal antibody, as compared to the control (Fig. 1C, 1D). The isolate was designated as CH/GDLZ/201801. After plaque purification and several passages, the viral titer reached 2.1 × 107 TCID50/mL, showing that the isolate was highly replicative in LMH cells.
Genome Size and Pylogenetic Analysis
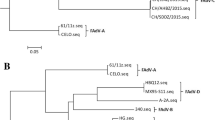
The genome of CH/GDLZ/201801 strain was 44,329 nt in length, with a G+C content of 58% and contained 44 open reading frames (ORFs) (Supplementary Table S1). The complete nucleotide sequences were deposited to the GenBank with accession number MK387061. Phylogenetic analysis based on the whole genome showed that strain CH/GDLZ/201801 clustered within FAdV-E with 97.3% identity to FAdV-8a TR59/Japan (KT862810) (Fig. 2A). The CH/GDLZ/201801 strain showed 96.7% identity to FAdV-8b HG/Canada (Grgić H et al. 2011), 96.5% identity to FAdV-8 HLJ-151129/China, 96.3% identity to FAdV-E FV211-16/Peru (Izquierdo-Lara et al. 2016) and 96.2% identity to FAdV-8b 764/United Kingdom.
Results of the phylogenetic analysis and recombination analysis of the strain CH/GDLZ/201801. A Phylogenetic tree of the nucleotide sequences of whole-genome. B SimPlot analysis was performed with the new isolate as the query sequence and FAdV-8a TR59, FAdV-8 HLJ-151129, FAdV-8b 764, FAdV-8b HG and FAdV-E FV211-16 as putative parental strains. Pale grey indicates the hexon, fiber and ORF19 CDS region, respectively. C Phylogenetic tree of the amino acid sequences of hexon. D Phylogenetic tree of the amino acid sequences of fiber. E Phylogenetic tree of the amino acid sequences of ORF19. Phylogenetic trees were constructed by using the neighbor-joining method (1000 bootstrap replicates). The recombinant isolate is labeled with red triangle.
Recombination Analysis
In order to detect possible recombination events within the new isolate CH/GDLZ/201801, genomic sequence analysis of the new isolate, FAdV-8a TR59, FAdV-8b HG, FAdV-8 HLJ-151129, FAdV-E FV211-16 and FAdV-8b 764 was carried out using the Simplot software. The result indicates that the recombination signal and the breakpoint of the new isolate are located at 21,021–21,961 nt, 31,561–32,741 nt and 34,721–37,821 nt. Furthermore, the recombination region consists of a hexon gene with 941 nt, a fiber gene with 1181 nt and a ORF19 with 3101 nt, and the region among three breakpoints is a recombinant region (Fig. 2B). The phylogenetic tree of the strains based on the amino acid sequences of hexon gene, fiber gene and ORF19 were also constructed and analyzed, demonstrating that the new isolate is actually a recombinant strain between FAdV-8a and FAdV-8b (Fig. 2C–2E).
Pathogenicity Assessment
The chickens in the control group did not show any clinical signs or necrotic lesions throughout the experiment. Three out of 20 chickens (15%) showed clinical depression, and 1 out of 20 animals (5%) died 7 dpi (Fig. 3). Necropsy revealed livers with pinpoint hemorrhages (Supplementary Figure S2) in some infected chickens.
Histopathological analysis showed that massive pathological damages were observed in various tissues of chickens in the infected group (Fig. 4). In the liver, basophilic inclusion bodies presented in hepatocytes with severe lymphocyte infiltration around vessels and fatty degeneration of hepatocytes (Fig. 4L). Lymphocytes greatly reduced and plenty of macrophages gathered in the spleen (Fig. 4M). In the lung, bronchioles structures were obliterated, blood stasis and dilation were observed in capillaries (Fig. 4N). Degeneration and structure disturbance were shown in the pancreas (Fig. 4Q). Abruption of intestinal villus was observed in the small intestine (Fig. 4R). A large number of exfoliated epithelial cells were observed in the proventriculus (Fig. 4S). A few necrotic and exfoliated epithelial cells were also observed in the gizzard (Fig. 4T). While, there was no significant histopathology damages presented in the tissues of chickens in control group (Fig. 4A–4J). In addition, immunohistochemical analysis showed that FAdV-8a antigens were detected in various tissues (Fig. 5), such as liver (Fig. 5L), spleen (Fig. 5M), lung (Fig. 5N), pancreas (Fig. 5Q), small intestine (Fig. 5R), proventriculus (Fig. 5S) and gizzard (Fig. 5T). No positive granules were detected in all the negative control of these organs (Fig. 5A–5J).
Hematoxylin and eosin (HE)-stained tissue sections. L Basophilic inclusion bodies in hepatocytes with severe lymphocyte infiltration and fatty degeneration. M Lymphocytes reduced and macrophages gathered in the spleen. N Bronchioles structures obliterated, blood stasis and dilation in lung capillaries. Q Degeneration and structure disturbance in the pancreas. R Abruption of intestinal villus in the intestine. S Exfoliated epithelial cells in the proventriculus. T necrotic and exfoliated epithelial cells in the gizzard. A–J A control chicken at 4 dpi. K–T A FAdV-8a-challenged chicken at 4 dpi.
Viral Shedding and Virus Distribution in Chickens Challenged with FAdV-8a
We explored the fecal virus shedding in FAdV-8a challenged chickens. As shown in Fig. 6A, the FAdV-8a DNA was detected by qPCR in fecal swabs collected from infected chickens from 2 to 12 dpi with 2 days interval, and peaked on 6 dpi. No FAdV-8a DNA was detected in the negative control chickens throughout the experiment. To examine the distribution of the FAdV-8a virus in different tissues in FAdV-8a-challenged chickens, three chickens from experimental group were necropsied at 4 dpi. As shown in Fig. 6B, the FAdV-8a DNA was detected in all collected samples. The results showed that the concentrations of the virus in all tissues were between 104 and 108 copies/μL, with the exceptions of concentrations higher than 109 copies/μL in livers. No FAdV-8a DNA was detected in the tissue samples from the control chickens. Taken together, these results demonstrate that FAdV-8a strain could be widely distributed in different tissues, but mainly concentrated in the livers of chicken.
Virus shedding in fecal swabs and various tissues of FAdV-8a-inoculated chickens. A Ct values and viral DNA shedding in fecal swabs after FAdV-8a or mock inoculation. B Virus distribution in various tissues of FAdV-8a-inoculated chickens at 4 dpi. Ct values and virus titer in various tissues after FAdV-8a inoculation. n = 3 chickens per group.
We identified virus particles in cell culture supernatants from LMH cells (Fig. 7A). To observe virus particles in vivo, the livers of chickens from each group were examined with TEM. No virus particles or pathological lesions were detected in the livers from the control chicken (Fig. 7B), and the latticed viral particles in the liver nuclei of an infected chicken with typical hexagonal structure of 70 nm in diameter and icosahedral particles indicated that FAdV-8a (CH/GDLZ/201801) strain could replicate in the livers of chicken (Fig. 7C). Taken together, these results show that the FAdV-8a strain could replicate and cause liver lesions in chickens.
Electron micrographs of FAdV-8a. A Electron micrographs of FAdV-8a from LMH cells. Typical icosahedral and non-enveloped particles (70 nm in diameter) were observed. B Electron micrographs of a control chicken liver at 4 dpi. C Electron micrographs of FAdV-8a (indicated by arrows) on a FAdV-8a-challenged chicken liver at 4 dpi.
Discussion
In recent years, outbreaks of FAdVs have been frequently reported worldwide, leading to huge economic losses to the poultry industry (Schachner et al. 2018). Since 2012, the clinical cases of IBH and HPS showed an increasing trend in China (Zhao et al. 2016; Li et al. 2016; Niu et al. 2018). In this study, a FAdV-8a strain CH/GDLZ/201801 was isolated from a case of IBH in Guangdong Province. The strain proved to be moderately pathogenic to 4-week-old chickens. This FAdV-8a strain can be further used for virological and serological assay development, as well as vaccine development.
A stable Leghorn male hepatoma cell line (LMH cellls) is commonly used to isolate FAdV (Kawaguchi et al. 1987). We attempted to isolate virus from FAdV-positive samples using LMH cells. After plaque purification and several passages, the viral titer reached 2.1 × 107 TCID50/mL, showing that CH/GDLZ/201801 strain was highly replicative in LMH cells. Consistent with our results, many studies showed that most FAdV strains can be propagated efficiently in LMH cells (Alexander et al. 1998; Ruan et al. 2018; Grgic et al. 2011). In addition, CPE was observed in infected LMH cells and was further verified by IFA with FAdV-8a fiber protein specific monoclonal antibody. The characteristic icosahedral particles of the purified FAdV-8a were observed by TEM.
In this study, the full genome of CH/GDLZ/201801 was 44,329 nucleotides in length compared to FAdV-8b HG (44,055 nt) and FAdV-8a TR59 (43,287 nt). The results of the sequence comparison and phylogenetic analysis show that the new isolate CH/GDLZ/201801 has the highest identity with FAdV-8a TR59 strain in terms of the complete genome, hexon gene and ORF19. Nevertheless, its fiber gene is closer to FAdV-8b HG rather than the other reference strains. These results suggest that the new isolate may be a recombinant strain of FAdV-8a and FAdV-8b. In addition, the recombination analysis further confirmed that the new isolate originates from a recombination between a FAdV-8a and a FAdV-8b strain.
In the current study, pathogenicity of FAdV-8a (CH/GDLZ/201801) was studied in SPF chickens. It indicated that three out of 20 chickens (15%) showed clinical depression, and one out of 20 animals (5%) died. Gross lesions and pathological changes by virus infection were obviously observed in the livers, and virus was found in all tissues and cloacal swabs of chickens with the highest viral copy numbers presented in the livers, which is similar to observations in other FAdV-8a strain infection (Xia et al. 2017). However, FAdV-8b isolates have no obvious pathogenicity in SPF chickens. For example, pathogenicity of FAdV-8b HG (Canadian strain) was studied in 10-day-old SPF chickens by Grgic et al. (2011). It showed that despite lack of clinical signs and pathological changes, virus was found in tissues and cloacal swabs of all birds with the highest viral copy numbers presented in the cecal tonsils. In addition, pathogenicity of FAdV-8b CH/SD/2015/09 (Chinese strain) was also studied in 5-week-old SPF chickens by Ruan et al. (2017). It showed that no clinical signs were observed in infected chickens, and no virus was detected in oral and cloacal swabs, and only viral DNA could be measured in kidneys.
In summary, we isolated a field strain of FAdV from a liver sample of a broiler chicken during a recent IBH outbreak. Phylogenetic analysis, based on the whole genome, showed that the new isolate manifests close relationship with FAdV-8a. Recombination analysis and phylogenetic analysis showed that the new isolate is a recombinant strain between FAdV-8a and FAdV-8b. Remarkably, three infected chicken showed clinical depression with a mortality of 5% in 4-week-old SPF chickens. Based on our present results, the novel Chinese isolate of FAdV-8a can be considered as moderately pathogenic to 4-week-old chickens. Nevertheless, further studies of the pathogenic mechanism and vaccine development of FAdV-8a are required to prevent and control the disease.
References
Alexander HS, Huber P, Cao J, Krell PJ, Nagy E (1998) Growth characteristics of fowl adenovirus type 8 in a chicken hepatoma cell line. J Virol Methods 74:9–14
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569
Choi KS, Kye SJ, Kim JY, Jeon WJ, Lee EK, Park KY, Sung HW (2012) Epidemiological investigation of outbreaks of fowl adenovirus infection in commercial chickens in Korea. Poult Sci 91:2502–2506
Domanska-Blicharz K, Tomczyk G, Smietanka K, Kozaczynski W, Minta Z (2011) Molecular characterization of fowl adenoviruses isolated from chickens with gizzard erosions. Poult Sci 90:983–989
Fender P, Boussaid A, Mezin P, Chroboczek J (2005) Synthesis, cellular localization, and quantification of penton-dodecahedron in serotype 3 adenovirus-infected cells. Virology 340:167–173
Gallina AM, Winterfield RW, Fadly AM (1973) Adenovirus infection and disease. II. Histopathology of natural and experimental disease. Avian Dis 17:343–353
Grafl B, Aigner F, Liebhart D, Marek A, Prokofieva I, Bachmeier J, Hess M (2012) Vertical transmission and clinical signs in broiler breeders and broilers experiencing adenoviral gizzard erosion. Avian Pathol 41:599–604
Grgic H, Philippe C, Ojkic D, Nagy E (2006) Study of vertical transmission of fowl adenoviruses. Can J Vet Res 70:230–233
Grgić H, Yang DH, Nagy E (2011) Pathogenicity and complete genome sequence of a fowl adenovirus serotype 8 isolate. Virus Res 156:91–97
Grimes TM, King DJ (1977) Effect of maternal antibody on experimental infections of chickens with a type-8 avian adenovirus. Avian Dis 21:12–97
Grimes TM, King DJ, Kleven SH, Fletcher OJ (1977) Involvement of a type-8 avian adenovirus in the etiology of inclusion body hepatitis. Avian Dis 21:26–38
Harrach B, Benkő M, Both GW, Brown M, Davison AJ, Echavarria M, Hess M, Jones MS, Kajon A, Lehmkuhl HD, Mautner V, Mittal SK, Wadell G (2011) Family adenoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the international committee on taxonomy of viruses. Elsevier, San Diego, pp 125–141
Hess M (2000) Detection and differentiation of avian adenoviruses: a review. Avian Pathol 29:195–206
Izquierdo-Lara R, Calderón K, Chumbe A, Montesinos R, Montalván Á, González AE, Icochea E, Fernández-Díaz M (2016) Complete genome sequence of fowl aviadenovirus serotype 8b isolated in South America. Genome Announc 4:e01174
Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T (1987) Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res 47:4460–4464
Li H, Wang J, Qiu L, Han Z, Liu S (2016) Fowl adenovirus species C serotype 4 is attributed to the emergence of hepatitis-hydropericardium syndrome in chickens in China. Infect Genet Evol 45:230–241
Lim TH, Lee HJ, Lee DH, Lee YN, Park JK, Youn HN, Kim MS, Youn HS, Lee JB, Park SY, Choi IS, Song CS (2011) Identification and virulence characterization of fowl adenoviruses in Korea. Avian Dis 55:554–560
Liu D, Zhang XB, Yan ZQ, Chen F, Ji J, Qin JP, Li HY, Lu JP, Xue Y, Liu JJ, Xie QM, Ma JY, Xue CY, Bee YZ (2013) Molecular characterization and phylogenetic analysis of infectious bursal disease viruses isolated from chicken in South China in 2011. Trop Anim Health Prod 45:1107–1112
Liu L, Qu Y, Wang T, Wang G, Wang F, Liu S (2014) Skin involvement in lymphomas caused by Marek’s disease virus infection in Silkie chickens. J Vet Diagn Investig 26:302–307
Lu Z, Zhang L, Wang N, Chen Y, Gao L, Wang Y, Gao H, Gao Y, Li K, Qi X, Wang X (2015) Naturally occurring reassortant infectious bursal disease virus in northern China. Virus Res 203:92–95
Marek A, Gunes A, Schulz E, Hess M (2010) Classification of fowl adenoviruses by use of phylogenetic analysis and high-resolution melting-curve analysis of the hexon L1 gene region. J Virol Methods 170:147–154
Mase M, Nakamura K, Minami F (2012) Fowl adenoviruses isolated from chickens with inclusion body hepatitis in Japan, 2009–2010. J Vet Med Sci 74:1087–1089
Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, Anson EL, Bolanos RA, Chou HH, Jordan CM, Halpern AL, Lonardi S, Beasley EM, Brandon RC, Chen L, Dunn PJ, Lai Z, Liang Y, Nusskern DR, Zhan M, Zhang Q, Zheng X, Rubin GM, Adams MD, Venter JC (2000) A whole-genome assembly of Drosophila. Science 287:2196–2204
Nicklin SA, Wu E, Nemerow GR, Baker AH (2005) The influence of adenovirus fiber structure and function on vector development for gene therapy. Mol Ther 12:384–393
Niczyporuk JS, Czekaj H (2018) A comparative pathogenicity analysis of two adenovirus strains, 1/A and 8a/E, isolated from poultry in Poland. Arch Virol 163:3005–3013
Niu Y, Sun Q, Zhang G, Sun W, Liu X, Xiao Y, Shang Y, Liu S (2017) Pathogenicity and immunosuppressive potential of fowl adenovirus in specific pathogen free chickens. Poult Sci 96:3885–3892
Niu Y, Sun Q, Zhang G, Sun W, Liu X, Xiao Y, Shang Y, Liu S (2018) Epidemiological investigation of outbreaks of fowl adenovirus infections in commercial chickens in China. Transbound Emerg Dis 65:e121–e126
Ojkic D, Martin E, Swinton J, Vaillancourt JP, Boulianne M, Gomis S (2008) Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathol 37:95–100
Pallister J, Wright PJ, Sheppard M (1996) A single gene encoding the fiber is responsible for variations in virulence in the fowl adenoviruses. J Virol 70:5115–5122
Reed LJ, Muench H (1938) A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497
Ruan SF, Zhao J, Ren YC, Feng JL, Zhang GZ (2017) Phylogenetic analyses of fowl adenoviruses (FAdV) isolated in China and pathogenicity of a FAdV-8 isolate. Avian Dis 61:353–357
Ruan SF, Zhao J, Yin XM, He ZR, Zhang GZ (2018) A subunit vaccine based on fiber-2 protein provides full protection against fowl adenovirus serotype 4 and induces quicker and stronger immune responses than an inactivated oil-emulsion vaccine. Infect Genet Evol 61:145–150
Russell WC (2009) Adenoviruses: update on structure and function. J Gen Virol 90:1–20
Schachner A, Matos M, Grafl B, Hess M (2018) Fowl adenovirus-induced diseases and strategies for their control—a review on the current global situation. Avian Pathol 47:111–126
Steer PA, Sandy JR, O’Rourke D, Scott PC, Browning GF, Noormohammadi AH (2015) Chronological analysis of gross and histological lesions induced by field strains of fowl adenovirus serotypes 1, 8b and 11 in one-day-old chickens. Avian Pathol 44:106–113
Toogood CI, Murali R, Burnett RM, Hay RT (1989) The adenovirus type 40 hexon: sequence, predicted structure and relationship to other adenovirus hexons. J Gen Virol 70:3203–3214
Wang P, Zhang J, Wang W, Li T, Liang G, Shao H, Gao W, Qin A, Ye J (2018) A novel monoclonal antibody efficiently blocks the infection of serotype 4 fowl adenovirus by targeting fiber-2. Vet Res 49:29
Winterfield RW, Fadly AM, Gallina AM (1973) Adenovirus infection and disease. I. Some characteristics of an isolate from chickens in Indiana. Avian Dis 17:334–342
Xia J, Yao KC, Liu YY, You GJ, Li SY, Liu P, Zhao Q, Wen RWY, Huang XB, Cao SJ, Han XF, Huang Y (2017) Isolation and molecular characterization of prevalent Fowl adenovirus strains in southwestern China during 2015–2016 for the development of a control strategy. Emerg Microbes Infect 6:e103
Zhang X, Zhong Y, Zhou Z, Liu Y, Zhang H, Chen F, Chen W, Xie Q (2016) Molecular characterization, phylogeny analysis and pathogenicity of a Muscovy duck adenovirus strain isolated in China in 2014. Virology 493:12–21
Zhao J, Zhong Q, Zhao Y, Hu YX, Zhang GZ (2016) Correction: pathogenicity and complete genome characterization of fowl adenoviruses isolated from chickens associated with inclusion body hepatitis and hydropericardium syndrome in China. PLoS ONE 11:e161744
Author information
Authors and Affiliations
Contributions
LC and LY performed the experiments and drafted the manuscript. QZ participated in the design of the study. PP and YD participated in the experiments and generated part of the data. YZ and CX analyzed the data. YC conceived the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
Animal experiments were performed strictly in accordance with the guidelines of Sun Yat-sen University Institutional Animal Care and Use Committee. The research was conducted in the compliance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, L., Yin, L., Peng, P. et al. Isolation and Characterization of A Novel Fowl Adenovirus Serotype 8a Strain from China. Virol. Sin. 35, 517–527 (2020). https://doi.org/10.1007/s12250-019-00172-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12250-019-00172-7