Abstract
Nowadays, food authentication is more and more required given its relevance in terms of quality and safety. The seafood market is heavily affected by mislabelling and fraudulent substitutions/adulterations, especially for processed food products such as canned food items, due to the loss of morphological features. This study aims to develop new assays based on DNA to identify fresh mackerel (Scomber spp.) and commercial products. A new primer pair was de novo designed on the 5S rRNA gene and non-transcribed spacer (NTS), identifying a DNA mini-barcoding region suitable for species identification of processed commercial products. Moreover, to offer a fast and low-cost analysis, a new assay based on recombinase polymerase amplification (RPA) was developed for the identification of fresh ‘Sgombro’ (Scomber scombrus) and ‘Lanzardo o Occhione’ (Scomber japonicus and Scomber colias), coupled with the lateral flow visualisation for the most expensive species (Scomber scombrus) identification. This innovative portable assay has great potential for supply chain traceability in the seafood market.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nowadays, food authentication has become a major concern for addressing quality and safety issues (Soon 2022). The seafood market is heavily affected by inadvertent or deliberate events of adulteration or species substitution (Silva & Hellberg 2021), especially for processed food products, such as canned food, due to the loss of morphological features (Xing et al. 2020). In Europe, seafood labelling is regulated by two legislations: regulations (EU) No.1169/2011 and (EU) No 1379/2013, according to which the label ought not to mislead consumers. The Regulation (EU) 1169/2011 includes general assessments regarding mandatory food information that has to be available and easily accessible (Art. 12, comma 1,3), but does not include the commercial denomination or scientific name of the fish species sold as processed products (Paolacci et al. 2021). This lack of information could be misleading for consumers and encourage misidentification processes and fraud. Canned seafood products such as tuna, salmon, anchovies and mackerel are the most widely consumed fish products across the world due to their practicality of storage and consumption (Mottola et al. 2022). The COVID-19 pandemic has increased the European consumption of canned fish, and the sales of these food items boomed during the lockdown period in Europe, particularly in Southern European nations such as Spain, France and Italy. The European Centre for the Promotion of Imports from developing countries (CBI) reported that canned tuna consumption went up by 12% from January to May 2020 compared to the same period in 2019, reaching a 38.6% increase in countries like Italy (https://www.cbi.eu/market-information/fish-seafood/canned-fish/market-potential). Although tuna is the bestselling canned product, mackerel (Scomber spp.) is an emerging alternative (conserved in brine, olive or vegetable oil) due to its (reduced o) lower costs and its increasing use in the seafood market (Mottola et al. 2022). From a taxonomic point of view, the genus of mackerel is composed offour different species, namely S. scombrus (Linnaeus, 1758), S. japonicus (Houttuyn, 1782), S. colias (Gmelin, 1789) and S. australasicus (Cuvier & Valenciennes, 1832). The Atlantic mackerel S. scombrus is more expensive than the other species due to the excellent properties of the meat (Infante et al. 2007). The average price for S. japonicus/colias in 2016 was 0.58 €/Kg while for S. scombrus was 1.24 €/Kg (Working Document to WGWIDE, 2017). Because the morphological features are removed during processing, identifying the species is difficult and fraudulent substitution with cheaper species increases. To fight fraud, DNA-based techniques are widely presented in the scientific literature (Böhme et al. 2019; Nehal et al. 2021; Barbuto et al. 2010; Shokralla et al. 2015; Frigerio et al. 2021a, b). DNA barcoding is the most used approach, especially for the seafood market, and the U.S. Food and Drug Administration (FDA) uses it to control and identify fish species for regulatory compliance (Yancy et al. 2008). The standard DNA barcode presented by Hebert in 2003 for animal identification was the 5’ end portion of the mitochondrial cytochrome c oxidase subunit I (COI). Nevertheless, due to the length of this region (650 bp), it cannot be used in highly processed products, where industrial treatment such as high temperature and pressure can lead to DNA fragmentation. For this reason, a DNA mini-barcoding approach (about 100–200 bp) is more suitable for this typology of products (Filonzi et al. 2021).
This study aims to develop new DNA-based assays to identify mackerel commercial products. To identify the species of canned mackerel products, a new primer pair was de novo designed on the 5S rRNA gene and non-transcribed spacer (NTS), identifying a DNA mini-barcoding region suitable for transformed and processed commercial products. 5S rRNA gene and non-transcribed spacer (NTS) werechosen because for mackerel identification the COI fragment is not able to provide an unambiguous identification at species level (Mottola et al. 2022).
Despite the fact that DNA mini-barcoding is efficient in analysing processed products such as canned mackerel, this technique requires a few days, higher costs and specialised laboratory personnel to obtain the results. To overcome this obstacle in this study, a new assay for fresh specimens was developed, based on recombinase polymerase amplification (RPA), for the identification of the most common mackerel species sold in Italy, ‘Sgombro’ (Scomber scombrus) and ‘Lanzardo o Occhione’ (Scomber japonicus and Scomber colias).
RPA in literature has been reported mainly in food pathogen detection tests such as Escherichia coli O157 (Zhao et al., 2022), Campylobacter jejuni (Geng et al. 2019), Salmonella (Hu et al. 2019), Listeria monocytogenes and Staphylococcus aureus (Guo et al. 2019) but thanks to the easiness of the assay it is a promising method also for species detection. RPA assay coupled with lateral flow provides a fast (less than an hour) and cheap (< 5 €) test for companies and consumers, reducing the time required to get the results and allowing it to be used also by non-specialised laboratory personnel.
Material and Methods
Specimen Collection
In this study, a total of 27 samples were collected (Table 1). Reference specimens (SCFEM_01-04) were sampled at the Milan fish market (Milan, Italy) and morphologically identified by the quality manager and veterinary surgeon, Dr Valerio Ranghieri, while the commercially canned specimens (SCFEM_05-27) were collected from Italian supermarkets coming from four different companies. Commercial samples were chosen from the most important Italian brands and considering three different conservation liquids(olive oil, vegetable oil andbrine).
Primer Design for DNA Mini-barcoding
Primer pairs for DNA mini-barcoding were newly designed in silico. The region 5S RNA gene and NTS was identified as one of the most variable markers to distinguish between all the Scomber species (Aranishi, 2005). All nucleotide sequences of the 5S rRNA gene and NTS (104 sequences) for Scomber spp. were obtained from NCBI Nucleotide and were aligned using ClustalW2 software (www.ebi.ac.uk/Tools/msa/clustalw2/). The most conserved regions were identified using Bioedit software and a primer pair specific for the genus Scomber spp. was de novo designed. 5S rRNA region was tested with Primer–Blast tool available from NCBI (www.ncbi.nlm.nih.gov/tools/primer-blast/) to verify the specificity to the Scomber genus.
Primer Design for RPA Analysis
Primer pairs for RPA were identified in silico and were designed on the 5S rRNA gene and NTS. Differently from PCR primers, RPA primers require a length of 30–35 nucleotides. In order to identify species-specific couples of primers, all nucleotide sequences of the 5S rRNA region for Scomber spp. were obtained from NCBI Nucleotide and primers were designed as shown in the previous paragraph but focusing on variable regions.
DNA Extraction
For all samples listed in Table 1, gDNA was obtained starting from 20 mg of tissue by using DNeasy Blood & Tissue Kit (QIAGEN, Germany). Canned specimens (SCFEM_05-27) were pre-treated in order to clean the tissue from the conservation liquid such as oil (vegetable and olive) or brine. The products conserved under brine were washed three times with a physiological solution (NaCl 0.7%) mixing overnight at 4 °C. Oil and lipids were removed by soaking in chloroform/methanol/water (1:2:0.8) mixing overnight at room temperature (Chapela et al. 2007). Purified gDNA was checked for concentration and purity by using a Qubit 4 Fluorometer and Qubit dsDNA HS Assay Kit (Invitrogen, Carlsbad, California, United States).
DNA Mini-barcoding
A standard PCR amplification was performed using PCR Mix Plus (A&A Biotechnology, Poland) following the manufacturer’s instructions in a 25-μL reaction containing 1 μL 10 mM of each primer and 3 μL of gDNA. PCR cycles consisted of an initial denaturation step for 5 min at 95 °C, followed by 35 cycles of denaturation (45 s at 95 °C), annealing (45 s at 50 °C) and extension (1 min at 72 °C), and, hence, a final extension at 72 °C for 7 min. The amplicon was visualised by electrophoresis on agarose gel using 1.5% agarose Tris–acetate-EDTA (TAE) gel. Purified amplicons were bidirectionally sequenced at Eurofins Genomics (Germany). After manual editing, primer removal and pairwise alignment, all the tested samples (Table 1) identities were assessed by adopting a standard comparison approach against the GenBank database with BLASTn (Altschul et al. 1990). Each barcode sequence was taxonomically assigned to the animal species with the nearest matches (maximum identity > 99% and query coverage of 100%).
RPA Assay
An RPA reaction mix (TwistAmp® Basic, England and Wales) was prepared in a total volume of 50 μL containing 2.5 μL of Magnesium Acetate (MgOAc, added at the end of mix preparation), 2.4 μL of 10 mM for each primer, 29.5 μL of rehydration buffer, 10.2 μL of sterile water and 3 μL of gDNA and tested on reference specimens (SCFEM_01-04).
Before proceeding with the amplification, the mixture was shaken vigorously to start the reaction. The amplification reaction consisted of 4 min at 39 °C, subsequently, it was further stirred and then put back at 39 °C for 20 min. Unlike DNA barcoding, RPA requires the purification of the amplicons prior to gel electrophoresis to guarantee better performance. The QIAquick Gel Extraction Kit (QIAGEN) was used for the purification of amplicons. Amplicons occurrence was assessed by electrophoresis on agarose gel using 1.5% agarose Tris–acetate-EDTA (TAE) gel.
Lateral Flow Assay
HybriDetect—Universal Lateral Flow Assay Kit (Milenia Biotec GmbH, Germany) with gold particles was used for lateral flow assay. The lateral flow strip is designed to develop qualitative or semi-quantitative rapid test systems. Primer forward was labelled with fluorescein-5,6-isothiocyanate (FICT) and primer reverse with biotin (BIO) by Eurofins Genomics (Germany). The samples were mixed with the solution supplied in the kit, and then the strip was placed into the solution. The DNA of interest, labeled with FITC and biotin, binded first to the gold-labeled FITC-specific antibodies in the sample application area of the strip. The gold complexes travelled through the membrane, driven by capillary forces. Only the DNA with the gold particles binded the test line with the immobilized biotin-ligand molecules, generating a grey-blue band. Unbound gold particles migrate over the control band and will be captued by species-specific antibodies.
This assay was only tested with S. scombrus primer pairs on reference specimens (SCFEM_01-04). The assay was performed with a modification of the company protocol, consisting in using only 3 μL of DNA for the visualisation on the lateral flow. For limit of detection (LOD) evaluation, the dilution of 1:10–1, 1:10–2, 1:10–3, 1:10–4 and 1:10–5 starting from the reference of S. scombrus was tested.
Results
DNA Mini-barcoding
The primer pair for DNA mini-barcoding was designed to be specific to the Scomber genus. To analyse highly processed products with expected degraded DNA, the primer pair amplifies a region of 151–160 base pairs. In Fig. 1 is described the sequence alignment for primer pair design and the primers’ sequences are described in Table 2. To evaluate a specific approach for Scomber genus, the most conserved region (indicated in green in Fig. 1) was identified for primer pair design.
Sequence alignment of the 5S rRNA gene and NTS from NCBI Nucleotide. The sequences displayed are representative of all the haplotypes generated by using FaBox (1.61) (https://users-birc.au.dk/palle/php/fabox/). The coloured nucleotide bases indicated a mismatch. On the top of the consensus sequence, at position 259 and 418 are indicated the designed primer. This alignment was obtained by Geneious Prime
DNA extraction was successful for all the samples with high DNA quality and good yield (i.e. 5.7–24.2 ng/μL). Reference samples (SCFEM_01-04) were analysed with the primer pair designed in this study with success. All the electropherograms obtained were high quality and allowed to uniquely identify the species. Therefore, for all the reference specimens, it was possible to identify the species, proving the ability of this marker to correctly identify all the species belonging to the Scomber genus.
All commercial samples (SCFEM_05-27) were successfully identified at the species level by Sanger sequencing, despite their processing stage (cooked at high temperature and under pressure, conserved in vegetable oil, olive oil and brine). Also for commercial samples, high-quality electropherograms were obtained. Amplicons obtained for all samples (SCFEM_01-27) are represented in Fig. 2.
RPA Assay
For RPA analysis, two primer pairs were designed on variable regions to be species-specific to S. scombrus (“Sgombro”) and S. japonicus/colias (“Lanzardo o Sgombro Occhione”), in order to distinguish these two groups (see Figure S.1 in supplementary information). The specificity was confirmed by developing in silico PCR through the software primer BLAST. The primer pairs amplify a product of 173–178 bp and are shown in Table 2.
Both the primer pairs were tested on the reference samples S. scombrus, S. colias, S. japonicus and S. australasicus (SCFEM_01-04). After the amplification process and amplicon purification, amplicons were visualised on an electrophoresis agarose gel. Results are shown in Fig. 3. Both primer pairs were specific only to target species.
These results allow us to distinguish S. japonicus and S. colias from S. scombrus which is the most expensive mackerel species, confirming the feasibility of the assay.
Lateral Flow Assay
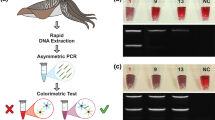
A lateral flow assay was developed only for the primer pair specific for S. scombrus, the most expensive species belonging to the Scomber genus. All the reference specimens (S. scombrus, S. colias, S. japonicus and S. australasicus) (SCFEM_01-04) and negative control were tested. The assay showed a band only for S. scombrus (Fig. 4a). In order to investigate the limit of detection (LOD), dilution series were created. The initial concentration of the S. scombrus sample was 16.6 ng/μL. Three microlitres of DNA dilution from 10–1 up to 10–5 was tested. Results of RPA reaction showed a high sensitivity of this assay, with a detection up to 0.0048 ng of total DNA (Fig. 4b).
a shows the results obtained from the lateral flow assay starting from RPA amplification. The upper band is the control band, the lower band shows the success of amplification. b shows the results of the dilution of the reference specimen of S. scombrus. Dilutions from 10–1 (4.98 ng of DNA) up to 10–5 (0.000498 ng of DNA) were tested
Discussion
The Misleading Labelling for Mackerel
Since it is not mandatory to provide the fish scientific name on labels for processed products, most of the commercial items purchased (i.e. 14) reported only the commercial name ‘Sgombro’ on their packaging. Based on Annex 1 of the Italian MiPAAF Decree dated September 22, 2017 (MiPAAF, DM 19,105, 2017), this term corresponds only to the S. scombrus species but it is commonly used in the market for all the Scomber species. In the other nine products analysed (see Table 1), only three provided the species name (S. colias) and the remaining six were labelled both as S. colias and S. japonicus which correspond to the commercial name ‘Lanzardo o Sgombro Occhione’. If we wanted to calculate a misidentification rate only considering the commercial name (“Sgombro”, corresponding to S.scombrus), 60% of canned products analysed in this study would result as mislabelled, a value higher than those reported in the scientific literature (Neo et al. 2022; Xing et al. 2020; Hu et al. 2018; Panprommin & Manosri 2022). Nevertheless, the product “Sgombro” sold canned, due to European Regulation not requiring the scientific name on the label for processed products, can be misinterpreted because it can be related to all mackerel species. Moreover, another labelling issue is related to the interchange of the two species S. colias and S. japonicus. Despite being two different species, as one inhabits the Atlantic/Mediterranean Sea (S. colias) and the other the Pacific Ocean (S. japonicus), both of these are accepted and sold under the same vernacular name ‘Lanzardo o Sgombro Occhione’ (Mottola et al. 2022). The old scientific name indeed was S. japonicus-colias, but only recent studies recognised the taxa as two allopatric species.
Canned and Fresh Mackerel Molecular Routine Analysis and Future Perspectives
Highly processed products such as canned fish can undergo transformation processes like high temperature and high pressure. These industrial processes could damage DNA, which can be fragmented and degraded. Literature shows that the DNA of canned fish products, such as tuna, sardines and mackerel is usually fragmented (Chapela et al. 2007; Pecoraro et al. 2020; Servusova & Piskata 2021). For this reason, a DNA mini-barcoding approach is required (Frigerio et al. 2021a, b; Roungchun et al. 2022). In this study, a couple of primers on the region 5S rRNA and NTS were de novo designed with success. The 5S rDNA consists of a 120 bp conserved region, but the length and sequence of the NTS may vary among species. Among nuclear markers, the 5S rRNA is the most interesting in taxonomic identification because of its unique structure making it a species specific gene in higher eukaryotes, including teleost fishes such as mackerel (Aranishi & Okimoto 2004). For this reason, the 5S rRNA region was chosen for the discrimination of Scomber species. Moreover, due to its lower length compared with the standard DNA barcoding region (about 650 bp), it can be useful to overcome the problem of processed and fragmented DNA. Using the primer pair designed in this study, it was possible to analyse and identify all commercial store-bought products, despite their processing stage (high temperature, under brine or oil conservation). Even if this technique can be successfully used for transformed products such as canned items, sometimes the fish market allows for short timeframes to complete the analysis. In fact, often it is not possible to store the fish for long periods (differently from other food sectors), especially when the product is to be sold fresh and not processed. Therefore, a standard DNA mini-barcoding approach is not suitable to meet the needs of this market. For this reason, in this study, we also wanted to develop an assay based on the RPA methodology for fresh mackerel detection, which allowed us to analyse a product in less than two hours and without the expensive instrumentation of a molecular biology laboratory. The lateral flows assay provides a visible result in less than 15 min with the visualisation of a band (in addition to the control band) in presence of the target species. This test is a ready-to-use, test strip based on lateral flow technology using gold particles.
In contrast to DNA mini-barcoding (see Table 3), the RPA assay is a specific analysis thataims to identify a specific species. In this study, we focused on differentiating the fresh products sold under the name of “Sgombro” (S. scombrus) and “Lanzardo” or “Sgombro occhione” (S. japonicus and S. colias) which are the most common in the seafood market. The former is a more expensive, higher quality fish compared to the latter. We had successfully developed and tested a specific couple of primers for S. scombrus and S. japonicus-S. colias for RPA assay with gel electrophoresis visualisation. In addition, in order to develop a rapid and cheap assay, the lateral flow assay for the couple of primers for S. scombrus, which is the most expensive species, was successfully developed and can detect very low quantity of DNA (0.0048 ng of total DNA). For companies, it could be a revolution in terms of supply chain traceability using a rapid and cheap kit that gives a result in less than two hours.
Excluding DNA barcoding and mini-barcoding techniques which allow universal analyses, other techniques besides RPA are presented in the literature for fish and mackerel authentication. A fast and easy tecnique similar to RPA is Loop-Mediated Isothermal Amplification (LAMP). An assay for fish detection has been reported to be fast (about 3 h) and with a limit of detection of 0.2 ng/μL (But et al. 2020). Prado et al. presented a real-time PCR method for mackerel detection with high sensitivity (up to 0.005 ng of DNA) (Prado et al. 2013). Although LAMP assay is faster and cheaper than real-time PCR, its sensitivity is low. RPA combine both the short timing and the high sensitivity (up to 0.0048 ng of total DNA), revealing to be the most promising methodology.For this reason, future studies will be focused on the development of the RPA assay with the lateral flow for the most common mackerel species, on further analysis on different specimen’s typology (such as canned products and multispecies products) and on assay validation before industrial scale-up and commercialization of a mock-up for companies A kit with this assay would be cheaper (< 5 €) and faster (less than an hour) than a laboratory test because it would not require expensive laboratory instrumentation and skilled technicians.
Conclusion
In this study, a DNA mini-barcoding and an RPA assay for the Scomber species identification were developed. The DNA mini-barcoding analysis enables the recognition of each species of Scomber (S. scombrus, S. colias, S. japonicus and S. australasicus). However, European legislation is still too permissive, as the determination of species on processed food labels is not yet mandatory. This can worsen fraud and mislabelling issues, which are already very common in the seafood sector. Furthermore, the development of a quick and cheap test, such as RPA and lateral flow assay, can be a huge change for companies, allowing for an economical control of the entire supply chain. Considering that this promising test is simple and easy to use, it would be used directly by consumers in the future, making them more aware of the products they buy and eat and protecting them from food fraud and mislabelling.
Data Availability
The data that support the findings of this study are available from the corresponding author J.F.
References
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Aranishi F (2005) Rapid PCR-RFLP method for discrimination of imported and domestic mackerel. Mar Biotechnol 7(6):571–575. https://doi.org/10.1007/s10126-004-4102-1
Aranishi F, Okimoto T (2004) PCR-based detection of allergenic mackerel ingredients in seafood. J Genet 83(2):193–195. https://doi.org/10.1007/BF02715826
Barbuto M, Galimberti, A, Ferri E, et al (2010) DNA barcoding reveals fraudulent substitutions in shark seafood products: the Italian case of “palombo”(Mustelus spp.). Food Res Int 43(1):376–381. https://doi.org/10.1016/j.foodres.2009.10.009
Böhme K, Calo-Mata P, Barros-Velázquez J, Ortea I (2019) Review of recent DNA-based methods for main food-authentication topics. J Agric Food Chem 67(14):3854–3864. https://doi.org/10.1021/acs.jafc.8b07016
But GWC, Wu HY et al (2020) Rapid detection of CITES-listed shark fin species by loop-mediated isothermal amplification assay with potential for field use. Sci Rep 10(1):1–14. https://doi.org/10.1038/s41598-020-61150-8
Chapela MJ, Sotelo CG, Pérez-Martín RI et al (2007) Comparison of DNA extraction methods from muscle of canned tuna for species identification. Food Control 18(10):1211–1215. https://doi.org/10.1016/j.foodcont.2006.07.016
Cuvier G, Valenciennes A (1832) Histoire naturelle des poissons. Tome huitième. Livre neuvième. Des Scombéroïdes. v. 8: i-xix + 5 pp. + 1-509, Pls. 209-245. [Cuvier authored pp. 1-470; Valenciennes 471-509. Date of 1831 on title page. i-xv + 1-375 in Strasbourg edition]
Filonzi L, Vaghi M, Ardenghi A et al (2021) Efficiency of DNA mini-barcoding to assess mislabeling in commercial fish products in Italy: an overview of the last decade. Foods 10(7):1449. https://doi.org/10.3390/foods10071449
Frigerio J, Marchesi C, Magoni C et al (2021) Application of DNA mini-barcoding and infrared spectroscopy for the authentication of the Italian product “bottarga.” LWT 139:110603. https://doi.org/10.1016/j.lwt.2020.110603
Frigerio J, Agostinetto G, Mezzasalma V et al (2021) DNA-based herbal teas’ authentication: an ITS2 and psbA-trnH multi-marker DNA metabarcoding approach. Plants 10(10):2120. https://doi.org/10.3390/plants10102120
Geng Y, Liu G, Liu L et al (2019) Real-time recombinase polymerase amplification assay for the rapid and sensitive detection of Campylobacter jejuni in food samples. J Microbiol Methods 157:31–36. https://doi.org/10.1016/j.mimet.2018.12.017
Gmelin JF (1789) Aves Anseres. Tome I. Pars II. In: Gmelin J.F. (Ed.) Caroli a Linnaei Systema Naturae per Regna Tria Naturae 1(2):501–1032
Guo Z, Chen Z, Liu X et al (2019) Detection of Listeria monocytogenes and Staphylococcus aureus via duplex recombinase polymerase amplification. J Food Saf 39(4):e12628. https://doi.org/10.1111/jfs.12628
Houttuyn M (1782) Beschryving van eenige Japanese visschen, en andere zee-schepzelen. Verh Holl Maatsch Wet Haarlem 20(2):311–350
Hu Y, Huang SY, Hanner et al (2018) Study of fish products in Metro Vancouver using DNA barcoding methods reveals fraudulent labeling. Food Control 94:38–47. https://doi.org/10.1016/j.foodcont.2018.06.023
Hu J, Huang R, Sun Y et al (2019) Sensitive and rapid visual detection of Salmonella Typhimurium in milk based on recombinase polymerase amplification with lateral flow dipsticks. J Microbiol Methods 158:25–32. https://doi.org/10.1016/j.mimet.2019.01.018
Infante C, Blanco E, Zuasti E et al (2007) Phylogenetic differentiation between Atlantic Scomber colias and Pacific Scomber japonicus based on nuclear DNA sequences. Genetica 130(1):1–8. https://doi.org/10.1007/s10709-006-0014-5
Linnaeus C (1758) Systema Naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Editio decima, reformata [10th revised edition], vol. 1: 824 pp. Laurentius Salvius: Holmiae
Mipaaf-Decreto Ministeriale n° (1910) 5 del 22 settembre 2017 - Denominazioni in lingua italiana delle specie ittiche di interesse commerciale. Available at: https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/11953#:~:text=di%20interesse%20commerciale-,Decreto%20Ministeriale%20n%C2%B019105%20del%2022%20settembre%202017%20%2D%20Denominazioni,tutte%20le%20Capitanerie%20di%20Porto. Accessed 29 April 2022
Mottola A, Piredda R, Catanese G et al (2022) Species authentication of canned mackerel: challenges in molecular identification and potential drivers of mislabelling. Food Control 137:108880. https://doi.org/10.1016/j.foodcont.2022.108880
Nehal N, Choudhary B, Nagpure A et al (2021) DNA barcoding: a modern age tool for detection of adulteration in food. Crit Rev Biotechnol 41(5):767–791. https://doi.org/10.1080/07388551.2021.1874279
Neo S, Kibat C, Wainwright BJ (2022) Seafood mislabelling in Singapore. Food Control 108821. https://doi.org/10.1016/j.foodcont.2022.108821
Panprommin D, Manosri R (2022) DNA barcoding as an approach for species traceability and labeling accuracy of fish fillet products in Thailand. Food Control 136:108895. https://doi.org/10.1016/j.foodcont.2022.108895
Paolacci S, Mendes R, Klapper R et al (2021) Labels on seafood products in different European countries and their compliance to EU legislation. Mar Policy 134:104810. https://doi.org/10.1016/j.marpol.2021.104810
Pecoraro C, Crobe V, Ferrari A (2020) Canning processes reduce the DNA-based traceability of commercial tropical tunas. Foods 9(10):1372. https://doi.org/10.3390/foods9101372
Prado M, Boix A, von Holst C (2013) Development of a real-time PCR method for the simultaneous detection of mackerel and horse mackerel. Food Control 34(1):19–23. https://doi.org/10.1016/j.foodcont.2013.04.007
Regulation (EU) No (1169), 2011 Of The European Parliament And Of The Council 2011, Available at: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32011R1169&from=IT. Accessed 29April 2022
Regulation (EU) No (1379), 2013 Of The European Parliament And Of The Council 2013, Available at: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32013R1379&from=IT. Accessed 29th April 2022
Roungchun JB, Tabb AM, Hellberg RS (2022) Identification of tuna species in raw and processed products using DNA mini-barcoding of the mitochondrial control region. Food Control 134:108752. https://doi.org/10.1016/j.foodcont.2021.108752
Servusova E, Piskata Z (2021) Identification of selected tuna species in commercial products. Molecules 26(4):1137. https://doi.org/10.3390/molecules26041137
Shokralla S, Hellberg RS, Handy SM et al (2015) A DNA mini-barcoding system for authentication of processed fish products. Sci Rep 5(1):1–11. https://doi.org/10.1038/srep15894
Silva AJ, Hellberg RS (2021) DNA-based techniques for seafood species authentication. Adv Food Nutr Res 95:207–255. https://doi.org/10.1016/bs.afnr.2020.09.001
Soon JM (2022) Food fraud countermeasures and consumers: a future agenda. In Future Foods 597–611. https://doi.org/10.1016/B978-0-323-91001-9.00027-X
The European and market potential for canned fish (2022). Available at: https://www.cbi.eu/market-information/fish-seafood/canned-fish/market-potential. Accessed 29 April 2022
Working Document to WGWIDE (2017) Available at: http://www.repositorio.ieo.es/e-ieo/bitstream/handle/10508/11071/Villamor_et_al.2017_WD%20WGWIDE_SColias.pdf?sequence=1&isAllowed=y. Accessed 29th April 2022
Xing RR, Hu RR, Han JX et al (2020) DNA barcoding and mini-barcoding in authenticating processed animal-derived food: a case study involving the Chinese market. Food Chem 309:125653. https://doi.org/10.1016/j.foodchem.2019.125653
Yancy HF, Fry SF, Randolph SC et al (2008) A protocol for validation of DNA-barcoding for the species identification of fish for FDA regulatory compliance. FDA Laboratory Information Bulletin
Zhao L, Wang J, Chen M, et al (2022) Development and application of recombinase polymerase amplification assays for rapid detection of Escherichia coli O157 in food. Food Analytical Methods 1-8 https://doi.org/10.1007/s12161-022-02250-1
Acknowledgements
We thank Dr Valerio Ranghieri, veterinarian surgeon and quality manager of the Milan Fish Market, who provided and morphologically recognised the SCFEM_01-04 samples. We thank Dr Paola Re for graphical support and Dr Filippo Bargero for English revision.
Funding
FEM2-Ambiente s.r.l., provided support in the form of a salary for authors J.F., V.M., T.M. and F.D.M.. The company only provided financial support in the form of research materials. No funding was received for this study.
Author information
Authors and Affiliations
Contributions
Conceptualization J.F, V.M., Methodology J.F., T.G., C.P., Validation J.F., T.G., V.M., Data Curation J.F., V.M., Writing—Original Draft J.F., V.M, T.G., Project administration V.M., J.F., M.L., Supervision Funding acquisition F.D.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of Interest
Jessica Frigerio declares that she has no conflict of interest. Tommaso Gorini declares that he has no conflict of interest. Cassandra Palumbo declares that she has no conflict of interest. Fabrizio De Mattia declares that he has no conflict of interest. Massimo Labra declares that he has no conflict of interest. Valerio Mezzasalma declares that he has no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Frigerio, J., Gorini, T., Palumbo, C. et al. A Fast and Simple DNA Mini-barcoding and RPA Assay Coupled with Lateral Flow Assay for Fresh and Canned Mackerel Authentication. Food Anal. Methods 16, 426–435 (2023). https://doi.org/10.1007/s12161-022-02429-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-022-02429-6