Abstract
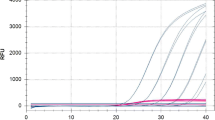
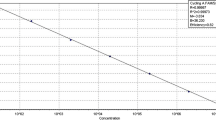
In this study, reverse transcription quantitative real-time PCR (RT-qPCR) assays using species-specific primers were developed for the quantification of the three major fungal species (Saccharomycopsis fibuligera, Rhizopus oryzae, and Monascus purpureus) during the traditional brewing of Hong Qu glutinous rice wine. Species-specific primers (Sfi-F/Sfi-R, Ro-F/Ro-R, and Mp-F/Mp-R) targeting the ITS-5.8S rRNA gene and the beta-tubulin gene sequences were designed and their specificity was evaluated by RT-qPCR with the total RNA from fungal species closely related to the traditional brewing of Hong Qu glutinous rice wine. Results of RT-qPCR using species-specific primers showed 100 % inclusivity (positive signal in the presence of target complementary DNA (cDNA)) and 100 % exclusivity (negative signals in the presence of non-target template). The specificity of each primer set was also tested when the target cDNA was mixed with non-target cDNA, and results showed that there was no significant difference (P > 0.05) in threshold cycle (Ct) values obtained from pure target cDNA and target cDNA mixed with non-target cDNA. The detection limits of the established qPCR assays were determined to be 101 gene copies/μL for S. fibuligera and 102 gene copies/μL for R. oryzae and M. purpureus, respectively. Good correlations were obtained for the tested species between 102 and 108 gene copies/μL by qPCR analysis. Finally, the established RT-qPCR assays were applied to quantify viable S. fibuligera, R. oryzae, and M. purpureus during the traditional brewing of Hong Qu glutinous rice wine. This study standardized the RT-qPCR assays for dominant fungi associated with the traditional brewing of Hong Qu glutinous rice wine, and it also proved the usefulness of RT-qPCR assays for the quantification of live cells in multi-species samples. Results would show great value in scientifically understanding of the brewing mechanism of Hong Qu glutinous rice wine.

Similar content being viewed by others
References
Andorrà I, Esteve-Zarzoso B, Guillamón JM, et al. (2010) Determination of viable wine yeast using DNA binding dyes and quantitative PCR. Int J Food Microbiol 144:257–262
Andorra I, Monteiro M, Esteve-Zarzoso B, et al. (2011) Analysis and direct quantification of Saccharomyces cerevisiae and Hanseniaspora guillermondii populations during alcoholic fermentation by fluorescence in situ hybridization, flow cytometry and quantitative PCR. Food Microbiol 28(8):1483–1491
Baldrian P, Větrovský T, Cajthaml T, et al. (2013) Estimation of fungal biomass in forest litter and soil. Fungal Ecol 6(1):1–11
Bramorski A, Christen P, Ramirez M, et al. (1998) Production of volatile compounds by the edible fungus Rhizopus oryzae during solid state cultivation on tropical agro-industrial substrates. Biotechnol Lett 20:359–362
Cawthorn DM, Witthuhn RC (2008) Selective PCR detection of viable Enterobacter sakazakii cells utilizing propidium monoazide or ethidium bromide monoazide. J Appl Microbiol 105:1178–1185
Chi ZM, Chi Z, Liu G, et al. (2009) Saccharomycopsis fibuligera and its applications in biotechnology. Biotechnol Adv 27:423–431
Christen P, Bramorski A, Revah S, et al. (2000) Characterization of volatile compounds produced by Rhizopus strains grown on agro-industrial solid wastes. Bioresour Technol 71:211–215
Du LQ, Tang C, Gu JQ, et al. (2005) Exploration to the esterification property of esterase produced by Monascus spp. China Brewing 142:23–24 (In Chinese)
Dung NTP, Rombouts FM, Nout MJR (2007) Characteristics of some traditional Vietnamese starch-based rice wine fermentation starters (men). LWT Food Sci Technol 40:130–135
Esteve-Zarzoso B, Belloch C, Uruburu F, et al. (1999) Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int J Syst Evol Microbiol 49:329–337
Feng XM, Larsen TO, Schnürer J (2007) Production of volatile compounds by Rhizopus oligosporus during soybean and barley tempeh fermentation. Int J Food Microbiol 113:133–141
Fey A, Eichler S, Flavier S, et al. (2004) Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl Environ Microbiol 70:3618–3623
Fittipaldi M, Nocker A, Codony F (2012) Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J Microbiol Methods 91:276–289
Fredlund E, Gidlund A, Olsen M, et al. (2008) Method evaluation of Fusarium DNA extraction from mycelia and wheat for downstream real-time PCR quantification and correlation to mycotoxin levels. J Microbiol Methods 73:33–40
Greppi A, Ferrocino I, La Storia A, et al. (2015) Monitoring of the microbiota of fermented sausages by culture independent rRNA-based approaches. Int J Food Microbiol 212:67–75
Hierro N, Esteve-Zarzoso B, Gonzalez A, et al. (2006) Real time quantitative PCR (qPCR) and reverse transcription-qPCR for detection and enumeration of total yeast in wine. Appl Environ Microbiol 72:7148–7155
Hierro N, Esteve-Zarzoso B, Mas A, et al. (2007) Monitoring of Saccharomyces and Hanseniaspora populations during alcoholic fermentation by real-time quantitative PCR. FEMS Yeast Res 7:1340–1349
Horváthová V, Slajsová K, Sturdík E (2004) Evaluation of the glucoamylase Glm from Saccharomycopsis fibuligera IFO 0111 in hydrolysing the corn starch. Biologia 59:361–365
Huang C-H, Shiu S-M, Wu M-T, et al. (2013) Monacolin K affects lipid metabolism through SIRT1/AMPK pathway in HepG2 cells. Arch Pharm Res 36:1541–1551
Kaberdin VR, Singh D, Lin CS (2011) Composition and conservation of the mRNA-degrading machinery in bacteria. J Biomed Sci 18(23):1–12
Kobayashi H, Oethinger M, Tuohy MJ, et al. (2009) Unsuitable distinction between viable and dead Staphylococcus aureus and Staphylococcus epidermis by ethidium bromide monoazide. Lett Appl Microbiol 48:633–638
Kubista M, Andrade JM, Bengtsson M, et al. (2006) The real-time polymerase chain reaction. Mol Asp Med 27:95–125
Lee C-L, Lin P-Y, Hsu Y-W, et al. (2015) Monascus-fermented monascin and ankaflavin improve the memory and learning ability in amyloid of total yeast in winetiond systematically for routine analysis of viable pop’s disease risk factors. J Funct Foods 18:387–399
Lee JL, Levin RE (2007) Quantification of total viable bacteria on fish fillets by using ethidium bromide monoazide real-time polymerase chain reaction. Int J Food Microbiol 118:312–317
Llanos Frutos R, Fernandez-Espinar MT, Querol A (2004) Identification of species of the genus Candida by analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Anton Leeuw Int J G 85:175–185
Lv XC, Huang ZQ, Zhang W, et al. (2012) Identification and characterization of filamentous fungi isolated from fermentation starters for Hong Qu glutinous rice wine brewing. J Gen Appl Microbiol 58(1):33–42
Lv XC, Huang XL, Zhang W, et al. (2013a) Yeast diversity of traditional alcohol fermentation starters for Hong Qu glutinous rice wine brewing, revealed by culture-dependent and culture-independent methods. Food Control 34(1):183–190
Lv XC, Huang RL, Chen F, et al. (2013b) Bacterial community dynamics during the traditional brewing of Wuyi Hong Qu glutinous rice wine as determined by culture-independent methods. Food Control 34(2):300–306
Lv XC, Cai QQ, Ke XX, et al. (2015a) Characterization of fungal community and dynamics during the traditional brewing of Wuyi Hong Qu glutinous rice wine by means of multiple culture-independent methods. Food Control 54:231–239
Lv XC, Chen ZC, Jia RB, et al. (2015b) Microbial community structure and dynamics during the traditional brewing of Fuzhou Hong Qu glutinous rice wine as determined by culture-dependent and culture-independent techniques. Food Control 57:216–224
Matsuda K, Tsuji H, Asahara T, et al. (2007) Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription-PCR. Appl Environ Microbiol 73:32–39
Nocker A, Cheung CY, Camper AK (2006) Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods 67(2):310–320
Nogva HK, Dromtorp S, Nissen H, et al. (2003) Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. Biotechniques 34(4):804–813
Park HG, Stamenova EK, Hong SC (2004) Phylogenetic relationships of Monascus species inferred from the ITS and the partial β-tubulin gene. Bot Bull Acad Sin 45:325–330
Rodríguez A, Rodríguez M, Luque MI, et al. (2012a) A comparative study of DNA extraction methods to be used in real-time PCR based quantification of ochratoxin A-producing molds in food products. Food Control 25:666–672
Rodríguez A, Rodríguez M, Martín A, et al. (2012b) Presence of ochratoxin A on the surface of dry-cured Iberian ham after initial fungal growth in the drying stage. Meat Sci 90:728–734
Rudi K, Moen B, Dromtorp SM, et al. (2005) Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl Environ Microbiol 71:1018–1024
Suanthie Y, Cousin MA, Woloshuk CP (2009) Multiplex real-time PCR for detection and quantification of mycotoxigenic Aspergillus, Penicillium and Fusarium. J Stored Prod Res 45:139–145
Taira J, Miyagi C, Aniya Y (2002) Dimerumic acid as an antioxidant from the mold, Monascus anka: the inhibition mechanisms against lipid peroxidation and hemeprotein-mediated oxidation. Biochem Pharmacol 63:1019–1026
Thanh VN, Mai LT, Tuan DA (2008) Microbial diversity of traditional Vietnamese alcohol fermentation starters (banh men) as determined by PCR mediated DGGE. Int J Food Microbiol 128:268–273
Varma M, Field R, Stinson M, et al. (2009) Quantitative real-time PCR analysis of total and propidium monoazide-resistant fecal indicator bacteria in wastewater. Water Res 43(19):4790–4801
Vendrame M, Iacumin L, Manzano M, et al. (2013) Use of propidium monoazide for the enumeration of viable Oenococcus oeni in must and wine by quantitative PCR. Food Microbiol 35:49–57
Vendrame M, Manzano M, Comi G, et al. (2014) Use of propidium monoazide for the enumeration of viable Brettanomyces bruxellensis in wine and beer by quantitative PCR. Food Microbiol 42:196–204
Wang CL, Shi DJ, Gong GL (2008) Microorganisms in Daqu: a starter culture of Chinese Maotai-flavor liquor. World J Microbiol Biotechnol 24:2183–2190
Xie GF, Li WJ, Lu J, et al. (2007) Isolation and identification of representative fungi from Shaoxing rice wine wheat qu using a polyphasic approach of culture-based and molecular-based methods. J I Brewing 113:272–279
Yan L, Zhang C, Dung L, et al. (2008) Development of a real-time PCR assay for the detection of Cladosporium fulvum in tomato leaves. J Appl Microbiol 104:1417–1424
Yang JH, Tseng YH, Lee YL, et al. (2006) Antioxidant properties of methanolic extracts from monascal rice. LWT Food Sci Technol 39:740–747
Yang X, Badoni M, Gill CO (2011) Use of propidium monoazide and quantitative PCR for differentiation of viable Escherichia coli from E. coli killed by mild or pasteurizing heat treatments. Food Microbiol 28:1478–1482
Zhu H, Qu F, Zhu LH (1993) Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res 21:5279–5280
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This work was financially supported by grants from the Project Funded by China Postdoctoral Science Foundation (2015M570549), Projects of Science and Technology of Fujian Education Dept., China (JA14107), and Scientific Research Start-up Funding of Fujian Agriculture and Forestry University (grant no. KXML2012A).
Conflict of Interest
Xu-Cong Lv declares that he has no conflict of interest. Rui-Bo Jia declares that he has no conflict of interest. Jing-Hao Chen declares that he has no conflict of interest. Wen-Bin Zhou declares that he has no conflict of interest. Yan Li declares that she has no conflict of interest. Bing-Xin Xu declares that she has no conflict of interest. Yi-Ting Liang declares that she has no conflict of interest. Bin Liu declares that he has no conflict of interest. Shao-Jun Chen declares that he has no conflict of interest. Yu-Ting Tian declares that he has no conflict of interest. Ping-Fan Rao declares that he has no conflict of interest. Li Ni declares that she has no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Not applicable in this study.
Additional information
Highlights
• Novel species-specific PCR primers for S. fibuligera, R. oryzae, and M. purpureus were developed.
• The specificity of the species-specific PCR primers was confirmed by RT-qPCR.
• The established assays were applied to the traditional brewing of Hong Qu glutinous rice wine.
Rights and permissions
About this article
Cite this article
Lv, XC., Jia, RB., Chen, JH. et al. Development of Reverse Transcription Quantitative Real-Time PCR (RT-qPCR) Assays for Monitoring Saccharomycopsis fibuligera, Rhizopus oryzae, and Monascus purpureus During the Traditional Brewing of Hong Qu Glutinous Rice Wine. Food Anal. Methods 10, 161–171 (2017). https://doi.org/10.1007/s12161-016-0565-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-016-0565-8