Abstract
Lignin is a cell wall polymer that reduces the enzymatic digestibility and conversion efficiency of lignocellulosic biomass to ethanol. Down-regulation of the hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT) gene reduces lignin content in alfalfa (Medicago sativa L.). The objective of this study was to evaluate HCT down-regulated alfalfa plants for their forage composition and agronomic performance in the greenhouse and under field conditions. Individual plants with two separate T0 HCT down-regulation events 3a and 30a were used to make crosses with a common male sterile line to generate T1 progenies (3A and 30A, respectively). The experimental design was a randomized complete block with four replications per line and ten individuals per replication. Two-row plots with full-sib progeny rows with (+) and without (−) the transgene were planted along with commercial cultivars used as checks. Most differential responses in forage quality and agronomic characteristics between full-sib down-regulated and non-down-regulated progenies were event-specific and found mainly between the HCT30A+ and HCT30A− progenies. Variation between HCT− and HCT+ alfalfa plants was observed in the HCT gene transcript levels, acid detergent lignin (ADL), relative feed value (RFV), and saccharification efficiency. Although differential responses in agronomic performance of field-grown HCT down-regulated alfalfa plants were identified for biomass yield and plant height, HCT+ and HCT− progenies had similar spring growth and fall dormancy. The reduction in lignin content of alfalfa plants via modification of HCT transcript levels increased forage quality and efficiency of sugar release of plants grown under greenhouse and field conditions.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
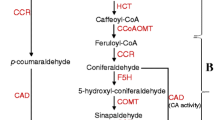
Alfalfa is a highly heterozygous, perennial forage legume. Legumes are important components of sustainable agricultural systems due to their capacity for symbiotic nitrogen fixation [1]. Perennial alfalfa can contribute to mitigate the negative impacts due to soil erosion, fertilizer run-off, and insecticide/pesticide leaching into water bodies [1,2,3]. Alfalfa is one of the major crops grown in the USA, is supported by established agricultural systems, and has a wide range of adaptability. The national alfalfa and forage alliance (NAFA) proposed the development of alfalfa as a dual-purpose crop in which the stems are used as biofuel feedstock and the leaves are used as high-value forage. Driving forces behind this effort include the generation and utilization of biomass-based biofuels to promote national energy independence from foreign-based fossil fuels [4] and to generate more carbon neutral energy resources. Lignin is a cell wall polymer that contributes to the recalcitrance of lignocellulosic biomass to enzymatic degradation and saccharification (sugar release from cell walls), thus reducing the processing efficiency of biomass-based bioethanol production [5, 6]. Lignin in dicots is composed of three monolignol units, namely p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S). The down-regulation of the lignin biosynthetic pathway gene hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT) was shown to reduce the abundance of G and S lignin monomers in alfalfa and increase the abundance of the H lignin monomer [7]. The G and S subunits are the two main monomers in alfalfa cell walls [7], and therefore, the reduced levels of these monomers result in an overall reduction in alfalfa lignin content. Hemicellulose and cellulose are also important components of plant cell walls, and, unlike lignin, these are partially digestible by ruminant animals [8]. Lignin content in alfalfa also impacts forage digestibility [9, 10] and can limit dry matter intake by animals [11] thus reducing animal gains. In addition to lignin quantity, the relative abundance of the lignin subunits also affects the efficiency by which the sugar molecules are released from the cell wall and the efficiency of biomass-based ethanol conversion [6, 12, 13].
Modification of lignin content in alfalfa was pursued using both traditional plant breeding approaches [9, 14] and transformation approaches by down-regulating enzymes in the lignin pathway [5, 7]. The outcomes from these approaches include increased sugar release from dried biomass of reduced-lignin alfalfa [7, 15, 16]. However, the gains in fermentable sugar release of lignin-modified plants are partially offset by reductions in plant yield, plant height, and field persistence in some of the lignin-modified plants [7, 9]. Crop yields can be reduced in plants with a significant reduction in lignin content, and in some cases, plants are more likely to lodge or have reduced persistence [17]. Positive effects on drought stress and disease responses were observed in reduced lignin alfalfa plants [16]. In poplar (Populas tremuloides Michx.), a negative correlation was identified between lignin content and plant growth rate [18].
At least ten genes from the lignin biosynthetic pathway have been well characterized thus far in flowering plants [19]. Six of these lignin biosynthetic genes were independently down-regulated in alfalfa and down-regulation of HCT in alfalfa lines resulted in the largest decrease in lignin content overall [5]. Assessing the agronomic performance and forage quality of HCT down-regulated ‘RegenSY’ alfalfa T0 plants in an environment beyond the greenhouse is critical to ascertain whether these variations are unique to each transformation event and determine if the gains in processing efficiency are maintained by plants when they are grown in the field. Thus, the objectives of this study were to evaluate the agronomic performance and forage composition of two HCT down-regulated events in alfalfa full-sib progenies grown under field conditions.
Materials and Methods
Plant Materials
HCT down-regulated alfalfa lines (T0) were generated using an antisense transgenic approach and an Agrobacterium-mediated transformation technique as previously described [7, 20] using RegenSY as the genetic background of the parent used for transformation. Briefly, the construct was generated from the M. truncatula full length HCT open reading frame (ORF) identified in the M. truncatula Gene Index (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=medicago). The construct included the PAL2 promoter which has preferential expression in vascular tissues [20]. Two T0 events (3a and 30a) were selected based on the strength of the HCT down-regulation for further evaluation of their agronomic performance. Event 3a had low levels of HCT transcripts while 30a had extremely low levels compared to the wild type as previously described [7]. Plants possessing the HCT down-regulated T0 events 3a and 30a were crossed by hand with separate plants from the male sterile line 208Ms-1 to generate the T1 progenies possessing the corresponding events, 3A and 30A, respectively. Additional crosses were made between 208Ms-1 and RegenSY and the empty vector line Ck-48. All crosses were performed in a greenhouse located in Ardmore, OK. Resulting seeds were scarified using sandpaper (2433 Emery Cloth Coarse, 3M, St. Paul, MN) prior to germination in cell flats containing Metro-Mix 350 Growing Media (Scotts-Sierra Hort. Products, Marysville, OH) and inoculated with Shinorhizobium meliloti (Royal Peat Alfalfa/Clover inoculum, Becker Underwood, Ames, IA). Segregation of the presence (+) or absence (−) of the transgene in the T1 progenies from each HCT event was identified using PCR primers previously described [7, 20]. The single copy number of the HCT transgene in the T0 plant and a subset of T1 progenies, as well as the 1:1 segregation ratio in the T1 progenies for the presence (+) and absence (−) of the transgene, were confirmed using southern blotting (DIG-High Prime DNA Labeling and Detection Starter Kit II, Roche Applied Science, Indianapolis, IN) of leaf DNA extracted as described in Xi et al. [21].
Field Plot Evaluations
The 8-week-old plants from the T1 seedlings segregating for the presence (+) and absence (−) of the transgene (HCT3A and HCT30A) and the corresponding checks were transplanted to a field in Ardmore, OK on 18 Oct, 2009 under USDA-APHIS permit 09-089-101-rm. The down-regulated (+) and non-down-regulated (−) full-sib T1 progeny lines for each event (HCT3A+/− and HCT30A+/−) were grown in adjacent one-row plots (one row of HCT+ plants next to one row of HCT− plants). The commercial alfalfa cultivars OK49 [22] and Bulldog505, a cultivar developed under grazing conditions (Athens Seed and Lawn, Inc), were used as checks and grown in one-row plots. The female parent 208Ms-1, the progenies from the cross 208Ms-1 × RegenSY (genetic background of the male parent), and the progenies from the cross 208Ms-1 × Ck-48 (empty vector transformed) were also included as controls. Soil at the study site was of the Normangee series (fine, smectitic, thermic Udertic Haplustalfs). The experimental design was a randomized complete block design with four replications and ten individual plants per replication in each plot, thus totaling 40 individuals evaluated for each line. Rows were spaced at 76.2 cm apart, with 38.1 cm between individual plants within each row. A border row of conventional lodging-tolerant alfalfa (45L133 line from Forage Genetics International, Nampa, ID) was used around the entire periphery of the field plot area. The field was irrigated, surveyed, and managed for weeds, insect pests, and diseases as needed throughout the growing season.
The aboveground biomass was harvested at 5-cm stubble height six times during the 2010 growing season on 24 Apr, 2 Jun, 7 Jul, 9 Aug, 13 Sep, and 29 Oct, seven times during the 2011 season on 14 Apr, 18 May, 14 Jun, 12 Jul, 4 Aug, 31 Aug, and 25 Oct, and four times during the 2012 season on 12 Apr, 10 May, 11 Jun, and 9 Jul. Biomass was harvested on a per plot basis and analyzed on a per plant basis (total plot biomass yield divided by the total number of existing plants per plot) to account for variations in plant persistence at each harvest during the multiple years after plant establishment. Harvested plant materials were devitalized (the harvested plant materials were allowed to reach terminal wilt) on-site based on permit requirements, air-dried in an oven at 50 °C for 96 h, and weighed to determine dry matter biomass yield. First harvest yield referred to as ‘spring growth’ was determined based on biomass yields from the first harvest in the spring of 2010, 2011, and 2012.
Agronomic characteristics including plant height, flowering traits, and lodging score were measured 1 to 2 days prior to each harvest in 2010, 2011, and 2012. Plant height was measured on the tallest stem of each plant to determine the natural plant height and used to generate a plot average. Percentage of canopy flowers within limits allowed by the permit was visually determined for each plot based on the percentage of the number of stems with flower(s). Lodging score was visually determined using a scale of 1 to 5 with 0.5 increments (1 represents a completely prostrate growth habit and 5 corresponds to the erect growth habit). The percentage of flowering plants at a given time was determined visually and used as an estimate of the time of flowering. Number of flowering plants was recorded before each harvest in 2011 within the permit guidelines.
Morphological and other agronomic traits evaluated include the leaf/stem ratio (LSR), fall dormancy, and plant persistence. LSR was measured from field-grown plants for harvests 1, 3, 4, 5, 6, and 7 in 2011, and for harvests 3 and 4 in 2012. Oven-dried samples were used to manually separate leaves and stems, and each fraction was subsequently weighed to determine the LSR. Fall dormancy evaluations were performed using plant height measurements based on a 5-cm increment scale for dormancy ratings starting after 5 to 8 weeks of growth from the final harvest in 2010 and 2011 (no 2012 data was collected because the field was terminated on 6 August, 2012 which was required prior to the expiration of the permit).
Forage quality estimates for acid detergent lignin (ADL), acid detergent fiber (ADF), neutral detergent fiber (NDF), crude protein, in vitro true dry matter digestibility (IVTDMD), and relative forage value (RFV) were determined for harvested samples using a near-infrared spectrometer Model 6500 (NIRSystems, Silver Springs, MD; now FOSS NIRSystems, Inc., Laurel, MD). Oven-dried biomass samples were ground using a 1-mm screen Wiley Mill (Thomas Model 4 Wiley Mill, Thomas Scientific, Swedesboro, NJ) and used to determine the ADL, ADF, NDF, crude protein, IVTDMD, and RFV with the alfalfa breeders’ prediction equation [23]. The forage quality values for the alfalfa samples obtained from the field were within the predictive range for the equation utilized and the data processing pipeline included a quality control step to confirm that no outliers with data outside the predictive ranges of the equation were detected based on both the global H value and the neighborhood H values. Forage quality analysis was determined for the aboveground tissues from all 2010 harvests and the first three harvests in 2011. Due to limitations in the percentage of flowering plants allowed in the permit, field-grown samples at the vegetative growth stage (13 nodal stages) were sampled on 28–29 Sept. 2011 for HCT3A+/− and HCT30A+/− and used for the HCT transcript analysis.
Greenhouse Evaluations
Lignin accumulation in the third, fifth, and seventh internodes (first internode refers to the one closest to the growing tip of stems) from clonally vegetative plants was estimated from plants grown in the greenhouse using vibrotome microtomy (Vibrotome 1000 Plus, Ted Pella, Inc., Redding, CA) for sample preparation and UV light microscopy (Nikon Microphot-FX, Melville, USA). The plants grown in the greenhouse correspond to clonal copies of the same plants grown in the field. Eighty-micro-meter-thick stem cross-sections of HCT3A+/− and HCT30A+/− lines were analyzed using UV microscopy at 10× magnification. Plant height, number of internodes, and average internode length were determined on HCT3A+/− and HCT30A+/− plants grown in a greenhouse in Ardmore, OK. Plants were vegetatively propagated and grown in the greenhouse under 24 °C day temperature and 18 °C night temperature with 16 h day-length (6:00 am to 22:00 h with supplementary lighting). The male parents (T0 plants) of both HCT down-regulated events (HCT3a and HCT30a) were included in the morphological measurements obtained from the greenhouse. The experimental design was a randomized complete block with six replications. Plants were cutback at 5 cm and height was measured after 5 weeks of regrowth on 16 Feb. 2012. The number of internodes was counted, stem length measured, and the average internode length determined.
HCT Transcripts and Saccharification Efficiency
Stem tissues from the 3rd to 6th internodes from the apical meristem were harvested on 5 July 2012 from the field-grown T1 (+) and (−) full-sib progenies of HCT3A and HCT30A plants at the 12 internode and early bloom stage of growth. The ground samples from alfalfa plants collected from the field and also used for the NIRS analysis were placed in a falcon tube and frozen in liquid nitrogen. Specifically, plants from four replications from two harvests in 2011 and two harvests in 2012 were analyzed. Greenhouse-grown plants were also harvested using the same protocol. Total RNA from samples was extracted using the RNeasy plant mini Kit (Qiagen Sciences, Germantown, MD, USA). RNA samples were treated with DNase I (Life Technologies, Grand Island, New York City, NY) following the manufacturer’s protocol. RNA concentrations were determined using a spectrophotometer ND 1000 (NanoDrop Technologies, Wilmington, DE) and the concentration was standardized to 300 ng/μl. RNA integrity was determined using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) prior to cDNA synthesis. cDNA was synthesized from the corresponding mRNA samples using Oligo(dT)20 primers and SuperScript III first strand synthesis system for RT-PCR (Life Technologies, Grand Island, New York City, NY) following the manufacturer’s protocol. cDNA samples were diluted 20-fold and used to determine the relative expression of HCT transcripts in HCT down-regulated and non-down-regulated samples via quantitative RT-PCR using the 7900HT System (Applied Biosystems-Life Technologies, Grand Island, New York City, NY). The M. truncatula ubiquitin (UBC) gene was used as an internal control and qPCR was performed using KiCqStart SYBR Green qPCR ReadyMix (Sigma-Aldrich, St. Louis, MO) following the manufacturer’s protocol.
Enzyme-released glucose (enzyme glucose), total glucose, and glucose release efficiency of stem tissues from the HCT+ and HCT− alfalfa progenies were determined using a phenol-sulfuric acid assay as previously described [5]. Briefly, dried and ground stem tissues were washed three times using chloroform/methanol (2:1 v/v) and water to obtain the cell wall residues (CWR). After drying overnight at 50 °C, 30 mg of CWR from each sample was collected in a glass tube for enzymatic sugar hydrolysis and total sugar hydrolysis using the procedure described in Chen and Dixon [5]. The sugar release was quantified based on the phenol sulfuric acid method and quantified using a Spectrophotometer [5].
Statistical Analysis
All phenotypic data were analyzed using the proc. MIXED procedure in SAS v9.2 (SAS Institute Inc., Cary, NC) considering year and transgene events as fixed effects and replication as a random effect after checking for normality and homoscedasticity of the data points. Mean comparison was performed at 5% of alpha error using Fischer’s LSD. For traits with significant line by year interaction, the data were analyzed separately for each year. Pearson’s correlation coefficients were determined for each phenotypic trait collected using proc. CORR also implemented in SAS.
Identification of the Transgene Insertion Site
Genomic DNA was extracted from T0 (HCT 3a and 30A) and T1 plant (HCT3A and HCT30A) events using MagAttract 96 DNA plant core kit (Qiagen Sciences, Germantown, MD, USA) following the manufacturer’s protocol. Specific PCR primers were designed using pCAMBIA2200 vector sequence for the left and right border (LB1: ATTTCACACAGGAAACAGCTATGA; RB1: ATGTGTGAGTAGTTCCCAGATAAG). One specific nested primer was designed for each left and right border sequence (LB2: GTTACCCAACTTAATCGCCTTG; RB2: TTAATTCGGCGTTAATTCAGTACA). Five arbitrary degenerate primers (AD1: NTCGASTWTSGWGTT; AD2: NGTCGASWGANAWGAA; AD3: WGTGNAGWANCANAGA; AD5: SSTGGSTANATWATWCT; AD6: CGSATSTCSAANAAWAT) were used for PCR reactions. ExTaq™ (Takara Biotechnology, Dalian, China) was used for PCR amplification in a 40-μl PCR reaction volume containing 25.4 μl water, 3 μl DNA (50 ng/μl), 1 μl gene specific primer (left border and right border specific primers were paired with each arbitrary degenerate primer separately) (10 μM), 3 μl AD primer (100 μM), 4 μl Takara ExTaq 10× buffer, 3.2 μl Takara dNTPs (2.5 mM), and 0.4 μl (5 units/ul) ExTaq enzyme. The second Tail-PCR was performed using specific primers nested within the first specific primer, by using 2 μl of 50× diluted PCR amplicons from the first reaction and PCR amplified using the same procedures and reagents as the first Tail-PCR as previously described in Cheng et al. [24]. Specific products from the HCT3A and HCT30A events were selected after screening the second Tail-PCR products on 1.5% agarose gel and purified using QIAquick PCR purification kit (Qiagen Sciences, Germantown, MD, USA). The purified PCR products were ligated and cloned using pGEM-T Easy vector (Promega Corporation, Madison, WI, USA). Colony PCR was performed using the nested primer and the AD primers. The colony PCR products with similar fragment sizes to that of the second Tail-PCR products were sequenced using the corresponding nested PCR primer that was used to amplify the fragment. DNA sequences were assembled into contigs using DNASTAR (DNASTAR, Inc., Madison, WI, USA) and only the contigs containing the nested primer and the vector sequence were retained. Sequences flanking the transgene insertion site for HCT3A and HCT30A events were cleaned by removing vector backbone from the assembled contigs and searched using Blastn against the M. truncatula genome sequence assembly (Mt4.0v1CDS) (available at: http://blast.jcvi.org/er-blast/index.cgi?project=mtbe) using WU-BLAST 2.0 to identify the putative chromosome of the insertion site.
Results
HCT Transcripts and Saccharification Efficiency
Differences in the relative expression of HCT transcripts between down-regulated and non-down-regulated full-sib progenies were identified for the two HCT30A and HCT3A events (Table 1). The HCT down-regulated parents (T0 HCT30a and T0 HCT3a) grown in the greenhouse and the corresponding HCT3A+ and HCT30A+ T1 progenies grown in the field had significantly lower relative expression levels of the HCT transcripts compared to the non-down-regulated progenies HCT3A− and HCT30A− (Table 1). Both the HCT30A+ and HCT3A+ T1 progenies had lower levels of expression in the greenhouse than in the field. The HCT30A− and HCT3A− have similar expression levels (1.0 and 1.1, respectively) which are higher than the corresponding T0 HCT30a and HCT3a down-regulated parental lines. Both the HCT30A+ and HCT3A+ plants have lower expression levels (0.27 and 0.16, respectively) than their corresponding HCT30A− and 3A− progenies (Table 1). The HCT30A+ plants grown in the greenhouse and sampled at the vegetative stage had lower lignin content at all three internodes evaluated compared to the HCT30A− plants (Fig. 1). After enzyme incubation, the HCT30A+ plants released higher amounts of glucose than the HCT30A− progenies, corresponding to 328 vs. 244 mg/g, respectively (Table 2). The total glucose was comparable for all lines evaluated. However, the glucose release efficiency of the HCT30A+ plants was higher (50%) than the corresponding HCT30A− full-sib progenies (Table 2).
Microscopic analysis of lignin deposition (shown as blue fluorescence) in stem cross sections (80 μm) under a UV microscope showing auto-fluorescence (10× magnification). The cross sections were taken before flowering from the 3rd, 5th, and 7th internodes counted from the apical meristems of non-down-regulated HCT30A− (a–c, respectively) and down-regulated HCT30A+ (d–f, respectively) in alfalfa
Agronomic and Morphological Traits
Agronomic characteristics evaluated in the field having a significant event effect based on the ANOVA include biomass yield, spring growth, plant height, flowering traits, lodging, and some forage quality traits (p value < 0.0001 to 0.05) (Table 3). The effect of year and the event by year interaction was trait-specific. Biomass yield, fall dormancy, leaf/stem ratio (LSR), relative feed value (RFV), in vitro true dry matter digestibility (IVTDMD), whole plant acid detergent lignin (ADL), whole plant acid detergent fiber (ADF), and whole plant neutral detergent fiber (NDF) did not have significant event by year interactions. The event by year interaction was significant (p > 0.05 and p > 0.01) for spring growth, plant height, canopy flowers, lodging score, and crude protein (CP) (Table 3). Differences in biomass yield between down-regulated and non-down-regulated full sib progenies were event-specific (Table 4). For example, HCT30A+ had lower biomass yields (254 g plant−1) compared to HCT30A− (340 g plant−1). In contrast, full-sib progenies from both HCT3A+ and HCT3A− had similar biomass yields. The yield values for these full-sib progenies were overall lower than the female parent 208Ms-1 (Table 4), which had the highest biomass yield at 423 g plant−1, followed by the commercial check OK49 (415 g plant−1). The HCT30A− had higher biomass yield than the RegenSY (318 g plant−1) and the Ck48 (313 g plant−1), but lower than the Bulldog505 (401 g plant−1).
The HCT30A+ plants had fewer flowering plants compared to the HCT30A− (Table 4). However, no differences were identified between flowering plants for the HCT3A− and the HCT3A+ event. The commercial checks OK49 and Bulldog505 and the female parent 208Ms-1 had a higher percentage of flowering plants compared to the HCT full-sib progenies. No differences in fall dormancy were observed between the HCT+ and HCT− progenies for both the 30A and 3A events. The leaf to stem ratio (LSR) had a significant effect due to event and year, but the interaction between line and year was not significant (Table 3). Specifically, HCT30A+ progenies had the highest LSR of all plants evaluated (Table 4), while the HCT30A− had a lower LSR value. The HCT3A− and HCT3A+ had similar LSR ratios, and these values were higher than the commercial checks and the female parent 208Ms-1 (Table 4).
Lignin Content and Other Forage Quality Characteristics
The HCT30A+ and HCT3A+ plants had, as expected, lower acid detergent lignin content (ADL) compared to their corresponding HCT30A− and HCT3A− full-sib progenies (Table 4). However, only the difference between the HCT30A+ and HCT30A− was statistically significant. The HCT30A+ plants had the lowest ADL (4.7% of dry matter, DM) of all the lines evaluated and this was significantly lower than the maternal line 208MS-1 and the check cultivars Bulldog595 and Ck48 (Table 4). In contrast, the values for ADF were similar for both the down-regulated and non-down-regulated progenies for both the HCT30A and the HCT3A plants. The whole plant NDF was higher for the commercial check OK49 but no differences overall were identified for NDF between both HCT+ and HCT− plants for both the 30A and 3A events. Significant differences between both the HCT30A+ and HCT30A− and the HCT3A+ and HCT3A− progenies were also identified for IVTDMD, with the HCT down-regulated lines having higher IVTDMD. The HCT30A+ also had significantly higher RFV than the HCT30A− plants. Although HCT3A+ had a slightly higher RFV value than the HCT3A− plants, these differences were not statistically significant (Table 4).
Due to the event by year interactions detected for spring growth, plant height, canopy flowering, LDG, and crude protein (Table 3), these data were analyzed separately for each year (Table 5). No differences in spring growth were identified in 2010 for all plants evaluated and no differences were observed in HCT3A+ and HCT3A− for subsequent years (2011 and 2012). However, differences in spring growth were identified between HCT30A+ and HCT30A− only in 2011. The spring re-growth in 2011 for all HCT plants evaluated was lower compared to the spring regrowth in 2010 and 2012. Differences in plant height between HCT30A+ and HCT30A− were identified in 2010 and 2012 but not in 2011. For the HCT3A+ and HCT3A−, no differences in plant height were observed between them (Table 5).
In the greenhouse, the T0 plants for both HCT3a and HCT30a were shorter than all other lines evaluated (Supplemental Table 1). Similar to the results from the field, the HCT30A+ plants were significantly shorter than the HCT30A− plants, while the HCT3A+ and HCT3A− plants had similar plant height. In regard to the number of internodes, the HCT30A+ had fewer internodes than HCT30A, while the HCT3A+ and HCT3A− had similar numbers of internodes (Supplemental Table 1). Meanwhile, the HCT30a T0 plants had the lowest number of internodes. The HCT+ plants for both 30A and 3A events had shorter internode lengths compared to the HCT− plants. The alfalfa cultivar Bulldog505 had a higher number of internodes than OK49 (Supplemental Table 1).
Variation in canopy flowering was detected for both the HCT30A and HCT3A between down-regulated and non-down-regulated progenies for 2010 and 2012, but not in 2011 (Table 5). No differences were detected for lodging or crude protein between HCT3A+ vs. HCT3A−, although differences in crude protein were identified between HCT30A+ and HCT30A− events in 2010 and 2011.
Lignin Deposition in Stems
Cross-sections of alfalfa stems revealed lower lignification in stem tissues from the HCT30A+ plants compared to HCT30A− plants (Fig. 1). The stem apex of HCT30A− had higher lignification in the vascular cambium (shown as blue fluorescence of lignified tissues between the vascular bundles, Fig. 1a) than the lignification of stems in the same internode of the HCT30A+ plants (Fig. 1d). The reduction in lignification of HCT30A+ plants was also observed in the fifth and seventh internodes (Fig. 1e, f) compared to the same internodes from HCT30A− (Fig. 1b, c). Differences in the lignification of stems between HCT3A+ and HCT3A− were not observed (data not shown). Despite the reduced accumulation of lignin content of HCT30A+ compared to HCT30A− (Table 4, Fig. 1), no differences were identified in the lodging score between them (Table 5). The lodging scores for the full-sib progenies were similar to those of RegenSY.
Trait Correlations
Biomass yield of alfalfa plants evaluated in the field was positively correlated with plant height, aboveground ADL, ADF, and NDF, and negatively correlated with LSR, CP, IVTDMD, and RFV (Supplemental Table 2). Lodging score was not significantly correlated with aboveground ADL, ADF, and NDF. The percentage of canopy flowering was positively correlated with aboveground ADL, ADF, and NDF and negatively correlated with LSR, CP, IVTDMD, and RFV.
HCT Transgene Insertion Site
Amplification of specific sequences for both HCT3A and HCT30A events were generated. For the HCT30A T0 event, sequences flanking the insertion site obtained from the right border-specific nester primer and primer AD3 primer (RB AD3) had the highest sequence similarity to genomic DNA corresponding to M. truncatula chromosome 7 (data not shown). For the HCT3A T0 event, the sequences flanking the insertion site were obtained using primer AD6 (RB AD6). The genomic sequence flanking the HCT3A T0 event had the highest similarity to DNA partially corresponding to chromosome 1 and chromosome 3 of M. truncatula (data not shown). These initial results suggest that the insertion sites for the T0 events HCT30A and HCT3A were located in different chromosomes.
Discussion
Value-added traits in agricultural systems require a predictable and stable expression of the transgene when grown under field conditions. Extensive evaluation and characterization of HCT in alfalfa were previously performed only in laboratory and greenhouse studies [5, 7, 16, 25]. Environmental conditions and stress factors impacting plants growing in the field can affect the expression of a transgene in ways that are not discernible during greenhouse studies. The HCT down-regulation of alfalfa was maintained in the T1 progenies generated from the original T0 plants crossed with the common female parent 208Ms-1. The HCT down-regulated alfalfa progenies had lower lignin content and greater forage quality in the field compared to the non-down-regulated progenies, thus confirming a similar effect of down-regulation both in the greenhouse and in the field.
Down-regulation of HCT resulted in lower lignin accumulation in stem tissues of plants from both 3A and 30A progenies similar to the stem lignin accumulation in the T0 parents previously described [7]. Field studies of regulated materials impose certain limitations on the percentage of flowering plants allowed, while plants evaluated under greenhouse conditions are not subject to these same restrictions. Lignin accumulation increases with plant maturity and therefore the differences in lignin content between HCT+ and HCT− lines should increase when plants are harvested at more mature stages. For bioethanol conversion, studies propose harvesting alfalfa at a more mature stage [26] as a strategy to maximize the benefit of lignin reduction due to HCT down-regulation and to reduce the number of harvests per year. The negative correlation between lignin content and saccharification efficiency in HCT down-regulated alfalfa plants shown here further exemplifies how lower lignin content can contribute to efficient bioethanol conversion [5, 6].
The event-specific lignin reduction in the T1 progenies (lower lignin in HCT30A+ compared to HCT3A+) was consistent with the lower accumulation of HCT transcripts in the corresponding T0 parental line HCT30a compared to the T0 parent HCT3a previously evaluated in the greenhouse [7]. The insertion site, promoter, and copy number of the transgene play a role in the growth and development of the resulting transformation events [27]. Both HCT3A and HCT30A refer to single copy insertions determined by the 1:1 segregation of the transgene in the progenies and confirmed with southern blot analysis. Therefore, it is possible that the difference in lignin accumulation between HCT30A and HCT3A events may be due to differences in the insertion sites of the transgene in the alfalfa genome [28, 29]. Indeed, our initial results based on flanking sequence of the insertion site suggest that events HCT3A and HCT30A are located on different chromosomes. Further analysis of the genetic content through targeted or whole-genome sequencing in these regions for different transformation events [30] beyond this study could provide additional insights aimed at further assessing the nature of these event-specific variations. Additionally, it is possible that variation between these events could be due to variation in methylation of the transgenes and/or their promoters and/or due to epigenetic variation as previously described [31, 32]. Hence, the importance of event sorting to identify the optimal transformation event for further evaluation as part of the trait-integration pipeline.
Improving the processing efficiency of alfalfa could have a significant economic impact [9, 10, 33] largely due to the predominance of alfalfa production as the 4th crop in acreage grown in the USA [34]. Increasing alfalfa forage quality has been an important aspect of alfalfa breeding programs for several decades [35, 36]. In this study, the HCT down-regulated alfalfa plants had lower lignin content and greater digestibility. These findings from field-grown plants are similar to previous studies in alfalfa in which lower lignin levels in plant cell walls correlate with higher amounts of digestible fibers [9, 33].
Lignin modification in alfalfa through genetic modification can impact other agronomic or morphological traits [7, 15, 16, 37]. In some cases, HCT down-regulation in alfalfa resulted in lower biomass yield, shorter plant height, delayed flowering, greater branching phenotype, shorter internode length, and greater disease and drought resistance [25]. These traits have relevance to practical field applications as they relate to alfalfa biomass yield and forage quality. Total biomass yield is considered an important trait for alfalfa production [38], while lignin content, rather than composition has a significant effect on alfalfa in situ digestibility [39]. Alfalfa plants with similar lignin content can have different biomass yield likely due to differences in the underlying genetic background and/or insertion site as proposed above. Commercial alfalfa cultivars with reduced lignin content developed through deactivation of enzymes in the lignin synthesis pathway or through conventional plant breeding have higher digestibility that conventional alfalfa varieties [40]. A study by Gallego-Giraldo et al. [25] showed that Arabidopsis plants down-regulated for lignin content resumed normal growth when the synthesis of salicylic acid was blocked. A previous study in alfalfa has reported a positive correlation between plant height and biomass yield [41]. We observed event-specific variation for plant height and biomass yield in HCT+ and HCT− progenies. A previous study reported a shorter internode length and fewer number of internodes for the original T0 HCT30A parent [16] that was also observed in the HCT30A+ progenies.
Overall, down-regulation of genes in the lignin biosynthetic pathway reduces lignin content in alfalfa and increases saccharification efficiency. The development of lignin down-regulated alfalfa plants with plant height and/or biomass yields more similar to the non-down-regulated lines could possibly be achieved through event sorting to identify those events with favorable insertion site locations and optimized gene expression levels. Additionally, integrating breeding approaches that involve crossing the reduced lignin plants with elite genotypes possessing desirable agronomic traits as part of a trait-development pipeline could be useful when considering the cost-benefit proposition of a future commercial product.
Abbreviations
- ADF:
-
acid detergent fiber
- ADL:
-
acid detergent lignin
- ANOVA:
-
analysis of variance
- BLAST:
-
basic local alignment search tool
- CP:
-
crude protein
- DOE:
-
United States Department of Energy
- HCT :
-
hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase
- IVTDMD:
-
in vitro true dry matter digestibility
- LSR:
-
Leaf/stem ratio
- NAFA:
-
National alfalfa and forage alliance
- NASS:
-
National Agriculture Statistics Service
- NDF:
-
neutral detergent fiber
- ORF:
-
open reading frame
- RFV:
-
relative feed value
- qRT-PCR:
-
reverse transcriptase–quantitative polymerase chain reaction
- Tail-PCR:
-
thermal asymmetric interlaced-polymerase chain reaction
- USDA:
-
United States Department of Agriculture
References
Russelle MP, Lamb JFS, Turyk NB, Shaw BH, Pearson B (2007) Managing nitrogen contaminated soils: benefits of N-2-fixing alfalfa. Agron J 99:738–746. https://doi.org/10.2134/agronj2006.0325.
Donner SD, Kucharik CJ (2008) Corn-based ethanol production compromises goal of reducing nitrogen export by the Mississippi River. Proc Natl Acad Sci U S A 105:4513–4518. https://doi.org/10.1073/pnas.0708300105
Syswerda SP, Basso B, Hamilton SK, Tausig JB, Robertson GP (2012) Long-term nitrate loss along an agricultural intensity gradient in the upper Midwest USA. Agric Ecosyst Environ 149:10–19. https://doi.org/10.1016/j.agee.2011.12.007
USDA/DOE 2005. Biomass as feedstock for a bioenergy and bioproducts industry: the technical feasibility of a billion-ton annual supply. Oak Ridge, TN. Available at: https://www.energy.gov/sites/prod/files/2014/04/f14/final_billionton_vision_report2.pdf
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25:759–761. https://doi.org/10.1038/nbt1316
Dien BS, Miller DJ, Hector RE, Dixon RA, Chen F, McCaslin M, Reisen P, Sarath G, Cotta MA (2011) Enhancing alfalfa conversion efficiencies for sugar recovery and ethanol production by altering lignin composition. Bioresour Technol 102:6479–6486. https://doi.org/10.1016/j.biortech.2011.03.022
Shadle G, Chen F, Reddy MSS, Jackson L, Nakashima J, Dixon RA (2007) Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68:1521–1529. https://doi.org/10.1016/j.phytochem.2007.03.022
Jung HG, Engels FM (2002) Alfalfa stem tissues: cell wall deposition, composition, and degradability. Crop Sci 42:524–534
Buxton DR, Casler MD (1993) Environmental and genetic factors affecting cell wall composition and digestibility. In: Jung HG et al (eds) Forage cell wall structure and digestibility. American Society of Agronomy, Madison, pp 685–714
Moore KJ, Jung HJG (2001) Lignin and fiber digestion. J Range Manag 54:420–430. https://doi.org/10.2307/4003113
Mertens DR (1994) Regulation of forage intake. In: Fahey GC Jr (ed) Forage quality, evaluation and utilization. American Society of Agronomy, Madison, pp 450–493
Davison BH, Drescher SR, Tuskan GA, Davis MF, Nghiem NP (2006) Variation of S/G ratio and lignin content in a Populus family influences the release of xylose by dilute acid hydrolysis. Appl Biochem Biotechnol 130:427–435. https://doi.org/10.1385/abab:130:1:427
Studer MH, DeMartini JD, Davis MF, Sykes RW, Davison B, Keller M, Tuskan GA, Wyman CE (2011) Lignin content in natural Populus variants affects sugar release. Proc Natl Acad Sci U S A 108:6300–6305. https://doi.org/10.1073/pnas.1009252108
Schwab PM, Lamb JFS, Sheaffer CC, Barnes DK (2005) Germplasm variability and environmental effects on stem cellulose and lignin concentration in alfalfa. J Agron Crop Sci 191:386–392
Casler MD, Buxton DR, Vogel KP (2002) Genetic modification of lignin concentration affects fitness of perennial herbaceous plants. Theor Appl Genet 104:127–131. https://doi.org/10.1007/s001220200015
Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA (2011a) Selective lignin down-regulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190:627–639. https://doi.org/10.1111/j.1469-8137.2010.03621.x
Pedersen JF, Vogel KP, Funnell DL (2005) Impact of reduced lignin on plant fitness. Crop Sci 45:812–819. https://doi.org/10.2135/cropsci2004.0155
Novaes E, Kirst M, Chiang V, Winter-Sederoff H, Sederoff R (2010) Lignin and biomass: a negative correlation for wood formation and lignin content in trees. Plant Physiol 154:555–561. https://doi.org/10.1104/pp.110.161281
Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44:337–363. https://doi.org/10.1146/annurev-genet-102209-163508
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001) Down-regulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88. https://doi.org/10.2307/3871154
Xi Y, Fu C, Ge Y, Xiao X, Wang Z-Y (2009) Agrobacterium-mediated transformation of switchgrass and inheritance of the transgenes. BioEnergy Research 4:275–283. https://doi.org/10.1007/s12155-009-9049-7
Caddel JL, Berberet RC, Shelton KT, Zarrabi AA (1992) Registration of 'OK 49′ alfalfa. Crop Sci 32:280
NIRSC (2007) Alfalfa breeder calibration equation: NIRS forage and feed testing consortium. Filename: ah50-3. Hillsboro, WI. Available at: https://www.nirsconsortium.org/optimizing-nirs-performance
Cheng X, Wen J, Tadege M, Ratet P, Mysore KS (2011) In: reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Ed: Andy Pereira. Methods Mol Biol 678:179–190. https://doi.org/10.1007/978-1-60761-682-5_13.
Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA (2011b) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci U S A 108:20814–20819. https://doi.org/10.1073/pnas.1117873108
Lamb JFS, Sheaffer CC, Samac DA (2003) Population density and harvest maturity effects on leaf and stem yield in alfalfa. Agron J 95:635–641
Ghahfarokhi MK, Dormiani K, Mohammadi A, Jafarpour F, Nasr-Esfahani MH (2017) Blastocyst formation rate and transgene expression are associated with gene insertion into safe and non-safe harbors in the cattle genome. Sci Rep 7:15432. https://doi.org/10.1038/s41598-017-15648-3
Day CD, Lee E, Kobayashi J, Holappa LD, Albert H, Ow DW (2000) Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev 14:2869–2880. https://doi.org/10.1101/gad.849600
Vain P, James VA, Worland B, Snape JW (2002) Transgene behaviour across two generations in a large random population of transgenic rice plants produced by particle bombardment. Theor Appl Genet 105:878–889. https://doi.org/10.1007/s00122-002-1039-5
Guttikonda SK, Marri P, Mammadov J, Ye L, Sea K, Richey K, Cruse J, Zhuang M, Gao Z, Evans C, Rounsley S, Kumatla SP (2016) Molecular characterization of transgenic events using next generation sequencing approach. PLoS One 11:e0149515
Kanazawa A, O'Dell M, Hellens RP (2007) Epigenetic inactivation of chalcone synthase-a transgene transcription in petunia leads to a reversion of the post-transcriptional gene silencing phenotype. Plant Cell Physiol 48:638–647. https://doi.org/10.1093/pcp/pcm028
Meyer P, Linn F, Heidmann I, Meyer H, Niedenhof I, Saedler H (1992) Endogenous and environmental factors influence 35S promoter methylation of a maize A1 gene construct in transgenic Petunia and its colour phenotype. Mol Gen Genet 231:345–352. https://doi.org/10.1007/bf00292701
Jung HG, Mertens DR, Payne AJ (1997) Correlation of acid detergent lignin and klason lignin with digestibility of forage dry matter and neutral detergent fiber. J Dairy Sci 80:1622–1628
USDA/NASS (U.S (2016) Department of agriculture, National Agricultural Statistics Service). Available at: https://www.nass.usda.gov/
Hill, R.R., J.S. Shenk, R.F. Barnes. 1988. Breeding for yield and quality. In: Alfalfa and alfalfa improvement. pp. 809–825. ASA-CSSA-SSSA. Madison, WI. USA
Riday H, Brummer EC, Moore KJ (2002) Heterosis of forage quality in alfalfa. Crop Sci 42:1088–1093
Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA (2008) Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): effects on lignin composition in specific cell types. New Phytol 179:738–750. https://doi.org/10.1111/j.1469-8137.2008.02502.x
Undersander, D.J (2001) Alfalfa yield and stand. University of Wisconsin. Extension Bulletin. Madison, WI. Available at: https://fyi.uwex.edu/forage/alfalfa-yield-and-stand/
Reddy MSS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci U S A 102:16573–16578
Sheaffer C, Undersander D 2015 New reduced lignin alfalfa varieties: a potential forage quality breakthrough. University of Minnesota Extension bulleting: Available at: https://www.extension.umn.edu/agriculture/forages/variety-selection-and-genetics/reduced-lignin-alfalfa-varieties/
Rimi F, Macolino S, Leinauer B, Lauriault LM, Ziliotto U (2010) Alfalfa yield and morphology of three fall-dormancy categories harvested at two phenological stages in a subtropical climate. Agron J 102:1578–1585. https://doi.org/10.2134/agronj2010.0193
Acknowledgements
This study was funded by the Oklahoma Bioenergy Center (OBC), the Oklahoma Center for the Advancement of Science and Technology (OCAST), and The Noble Research Institute. We thank Mark McCaslin from Forage Genetics International, Inc. for providing the 208Ms-1 alfalfa genotype used for crossing and 45L133 alfalfa genotype used for the border rows. We appreciate assistance from Yuanhong Han to search the HCT sequences against the M. truncatula and alfalfa databases, and Christy Motes for assistance with the field trials. We also thank Dennis Walker from the Forage Analysis Core Facility for processing the alfalfa samples for forage quality and suggestions from anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplemental Table 1
(DOCX 14 kb)
Supplemental Table 2
(DOCX 14 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bhattarai, K., Rajasekar, S., Dixon, R.A. et al. Agronomic Performance and Lignin Content of HCT Down-Regulated Alfalfa (Medicago sativa L.). Bioenerg. Res. 11, 505–515 (2018). https://doi.org/10.1007/s12155-018-9911-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-018-9911-6