Abstract
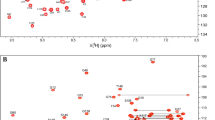
Human K-Ras protein, which is a member of the GTPase Ras family, hydrolyzes GTP to GDP and concomitantly converts from its active to its inactive state. It is a key oncoprotein, because several mutations, particularly those at residue position 12, occur with a high frequency in a wide range of human cancers. The K-Ras protein is therefore an important target for developing therapeutic anti-cancer agents. In this work we report the almost complete sequence-specific resonance assignments of wild-type and the oncogenic G12C and G12D mutants in the GTP-complexed active forms, including the functionally important Switch I and Switch II regions. These assignments serve as the basis for a comprehensive functional dynamics study of wild-type K-Ras and its G12 mutants.





Similar content being viewed by others
Data Availability
BMRB database (https://bmrb.io): access codes 52,021 (WT-GTP, 283 K), 52,023 (G12D-GTP, 283 K), and 52,024 (G21C-GTP, 288 K).
References
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Diercks T, Coles M, Kessler H (1999) An efficient strategy for assignment of cross-peaks in 3D heteronuclear NOESY experiments. J Biomol NMR 15:177–180
Ellis CA, Clark G (2000) The importance of being K-Ras. Cell Signal 12:425–434
Grzesiek S, Bax A (1993) Amino acid type determination in the sequential assignment procedure of uniformly 13 C/15 N-enriched proteins. J Biomol NMR 3:185–204
Hansen AL, Xiang X, Yuan C, Bruschweiler-Li L, Bruschweiler R (2023) Excited-state observation of active K-Ras reveals differential structural dynamics of wild-type versus oncogenic G12D and G12C mutants. Nat. Mol. & Struct. Biology (published August 28, 2023, https://doi.org/10.1038/s41594-023-01070-z)
Hunter JC, Gurbani D, Ficarro SB, Carrasco MA, Lim SM, Choi HG, Xie T, Marto JA, Chen Z, Gray NS, Westover KD (2014) In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C. Proc Natl Acad Sci USA 111:8895–8900
Hyberts SG, Takeuchi K, Wagner G (2010) Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J Am Chem Soc 132:2145–2147
Johnson BA, Blevins RA (1994) NMR view: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4:603–614
Kay LE, Keifer P, Saarinen T (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc 114: 10663–10665
Maciejewski MW, Schuyler AD, Gryk MR, Moraru II, Romero PR, Ulrich EL, Eghbalnia HR, Livny M, Delaglio F, Hoch JC (2017) NMRbox: a resource for Biomolecular NMR computation. Biophys J 112:1529–1534
Marcus K, Mattos C (2015) Direct Attack on RAS: Intramolecular Communication and Mutation-Specific effects. Clin Cancer Res 21:1810–1818
Marsh JA, Singh VK, Jia Z, Forman-Kay JD (2006) Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci 15:2795–2804
Menyhard DK, Palfy G, Orgovan Z, Vida I, Keseru GM, Perczel A (2020) Structural impact of GTP binding on downstream KR S signaling. Chem Sci 11:9272–9289
Palfy G, Menyhard DK, Akontz-Kiss H, Vida I, Batta G, Toke O, Perczel A (2022) The importance of Mg2+-Free state in Nucleotide Exchange of oncogenic K-Ras mutants. Chem Eur J 28:e202201449
Palfy G, Vida I, Perczel A (2020) 1H, 15 N backbone assignment and comparative analysis of the wild type and G12C, G12D, G12V mutants of K-Ras bound to GDP at physiological pH. Biomol NMR Assign 14:1–7
Palmer AG, Cavanagh J, Wright PE, Rance M (1991) Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J Magn Reson 93:151–170
Prior IA, Hood FE, Hartley JL (2020) The frequency of Ras mutations in cancer. Cancer Res 80(14):2969–2974
Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progress in NMR Spectroscopy 34:93–158
Sharma AK, Lee SJ, Rigby AC, Townson SA (2018) NMR 1H, 13 C, 15 N backbone and 13 C side chain resonance assignment of the G12C mutant of human K-Ras bound to GDP. Biomol NMR Assign 12:269–272
Showalter SA, Bruschweiler-Li L, Johnson E, Zhang F, Brüschweiler R (2008) Quantitative lid dynamics of MDM2 reveals differential ligand binding modes of the p53-binding cleft. J Am Chem Soc 130:6472–6478
Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW (2012) Discovery of Small molecules that bind to K-Ras and inhibit sos-mediated activation. Angew Chem Int Ed Engl 51(25):6140–6143
Vo U, Embrey KJ, Breeze AL, Golovanov AP (2013) 1H, 13 C and 15 N resonance assignment for the human K-Ras at physiological pH. Biomol NMR Assign 7:215–219
Xie M, Yu L, Bruschweiler-Li L, Xiang X, Hansen AL, Bruschweiler R (2019) Functional protein dynamics on uncharted time scales detected by nanoparticle-assisted NMR spin relaxation. Sci Adv 5(8):eaax5560
Ying J, Delaglio F, Torchia DA, Bax A (2017) Sparse multidimensional iterative lineshape-enhanced (SMILE) Reconstruction of both non-uniformly sampled and conventional NMR data. J Biomol NMR 68:101–118
Acknowledgements
This work was supported by the U.S. National Science Foundation (MCB-2103637). All NMR experiments were performed at the CCIC NMR facility at The Ohio State University.
Funding
U.S. National Science Foundation (MCB-2103637).
Author information
Authors and Affiliations
Contributions
L. B.-L. and R. B. conceived and designed the project. L. B.-L. prepared all K-Ras samples. C. Y. performed and analyzed all resonance assignment experiments. A.L.H. assisted in data collection. All authors contributed to the manuscript writing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, C., Hansen, A.L., Bruschweiler-Li, L. et al. NMR 1H, 13C, 15N backbone resonance assignments of wild-type human K-Ras and its oncogenic mutants G12D and G12C bound to GTP. Biomol NMR Assign (2023). https://doi.org/10.1007/s12104-023-10162-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12104-023-10162-2