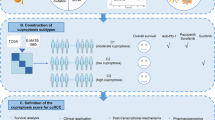
Abstract
Background
Cuproptosis, as a unique modality of regulated cell death, requires the involvement of ubiquitin-binding enzyme UBE2D2. However, the prognostic and immunotherapeutic values of UBE2D2 in pan-cancer remain largely unknown.
Methods
Using UCSC Xena, TIMER, Clinical Proteomic Tumor Analysis Consortium (CPTAC), and Human Protein Atlas (HPA) databases, we aimed to explore the differential expression pattern of UBE2D2 across multiple cancer types and to evaluate its association with patient prognosis, clinical features, and genetic variations. The association between UBE2D2 and immunotherapy response was assessed by gene set enrichment analysis, tumor microenvironment, immune gene co-expression and drug half maximal inhibitory concentration (IC50) analysis.
Results
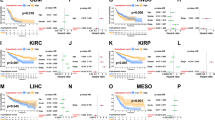
The mRNA and protein levels of UBE2D2 were markedly elevated in most cancer types, and UBE2D2 exhibited prognostic significance in liver hepatocellular carcinoma (LIHC), kidney chromophobe (KICH), uveal melanomas (UVM), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), and kidney renal papillary cell carcinoma (KIRP). UBE2D2 expression was correlated with clinical features, tumor mutation burden, microsatellite instability, and anti-tumor drug resistance in several tumor types. Gene enrichment analysis showed that UBE2D2 was significantly associated with immune-related pathways. The expression level of UBE2D2 was correlated with immune cell infiltration, including CD4 + T cells、Macrophages M2、CD8 + T cells in pan-cancer. PDCD1, CD274 and CTLA4 expression levels were positively correlated with UBE2D2 level in multiple cancers.
Conclusions
We comprehensively investigated the potential value of UBE2D2 as a prognostic and immunotherapeutic predictor for pan-cancer, providing a novel insight for cancer immunotherapy.










Similar content being viewed by others
Data availability
The data sets analyzed during the current study are available in the UCSC Xena database (http://xena.ucsc.edu/), CPTAC database (https://pdc.cancer.gov/pdc/), Human Protein Atlas database (http://www.proteinatlas.org/), CellMiner database (https://discover.nci.nih.gov/cellminer/home.do), and RCSB PDB database (https://www.rcsb.org/). Other data may be obtained from the corresponding author upon reasonable request.
Abbreviations
- ACC:
-
Adrenocortical carcinoma
- BLCA:
-
Bladder urothelial carcinoma
- BRCA:
-
Breast carcinoma
- CESC:
-
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL:
-
Cholangiocarcinoma
- CI:
-
Confidence interval
- COAD:
-
Colon adenocarcinoma
- CPTAC:
-
Clinical Proteomic Tumor Analysis Consortium
- DFI:
-
Disease-free interval
- DLBC:
-
Lymphoid neoplasm diffuse large B-cell lymphoma
- DSS:
-
Disease-specific survival
- ESCA:
-
Esophageal carcinoma
- GBM:
-
Glioblastoma multiforme
- GSEA:
-
Gene Set Enrichment Analysis
- GTEx:
-
Genote-Tissue Expression
- HNSC:
-
Head and neck squamous cell carcinoma
- HPA:
-
Human Protein Atlas
- HR:
-
Hazard ratio
- IC50:
-
Half maximal inhibitory concentration
- ICIs:
-
Immune checkpoint inhibitors
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KICH:
-
Kidney chromophobe
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- LAML:
-
Acute myeloid leukemia
- LGG:
-
Brain lower grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MESO:
-
Mesothelioma
- MSI:
-
Microsatellite instability
- NK:
-
Natural killer
- OS:
-
Overall survival
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PCPG:
-
Pheochromocytoma and paraganglioma
- PDB:
-
Protein Data Bank
- PFI:
-
Progression-free interval
- PRAD:
-
Prostate adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- SARC:
-
Sarcoma
- SKCM:
-
Skin cutaneous melanoma
- STAD:
-
Stomach adenocarcinoma
- TCA:
-
Tricarboxylic acid
- TCGA:
-
The Carcinoma Genome Atlas
- TGCT:
-
Testicular germ cell tumors
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- TIDE:
-
Tumor immune dysfunction and exclusion
- TMB:
-
Tumor mutational burden
- TME:
-
Tumor microenvironment
- UCEC:
-
Uterine corpus endometrial carcinoma
- UCS:
-
Uterine carcinosarcoma
- UVM:
-
Uveal melanomas
References
Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27.
Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–61.
Li SR, Bu LL, Cai L. Cuproptosis: lipoylated TCA cycle proteins-mediated novel cell death pathway. Signal Transduct Target Ther. 2022;7:158.
Cui L, Gouw AM, LaGory EL, Guo S, Attarwala N, Tang Y, et al. Mitochondrial copper depletion suppresses triple-negative breast cancer in mice. Nat Biotechnol. 2021;39:357–67.
Ramchandani D, Berisa M, Tavarez DA, Li Z, Miele M, Bai Y, et al. Copper depletion modulates mitochondrial oxidative phosphorylation to impair triple negative breast cancer metastasis. Nat Commun. 2021;12:7311.
Liao Y, Zhao J, Bulek K, Tang F, Chen X, Cai G, et al. Inflammation mobilizes copper metabolism to promote colon tumorigenesis via an IL-17-STEAP4-XIAP axis. Nat Commun. 2020;11:900.
Wang J, Qin D, Tao Z, Wang B, Xie Y, Wang Y, et al. Identification of cuproptosis-related subtypes, construction of a prognosis model, and tumor microenvironment landscape in gastric cancer. Front Immunol. 2022;13:1056932.
Michalczyk K, Cymbaluk-Płoska A. The role of zinc and copper in gynecological malignancies. Nutrients. 2020;12:3732.
Kazi Tani LS, Gourlan AT, Dennouni-Medjati N, Telouk P, Dali-Sahi M, Harek Y, et al. Copper isotopes and copper to zinc ratio as possible biomarkers for thyroid cancer. Front Med. 2021;8: 698167.
Ge EJ, Bush AI, Casini A, Cobine PA, Cross JR, DeNicola GM, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer. 2022;22:102–13.
Huang Y, Yin D, Wu L. Identification of cuproptosis-related subtypes and development of a prognostic signature in colorectal cancer. Sci Rep. 2022;12:17348.
Cockram PE, Kist M, Prakash S, Chen SH, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021;28:591–605.
Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–43.
Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8.
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–14.
Edwards NJ, Oberti M, Thangudu RR, Cai S, McGarvey PB, Jacob S, et al. The CPTAC data portal: a resource for cancer proteomics research. J Proteome Res. 2015;14:2707–13.
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27.
Colwill K, Gräslund S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods. 2011;8:551–8.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1:274–8.
Yu G, Wang LG, Han Y, He QY. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Schober P, Boer C, Schwarte LA. Correlation coefficients: appropriate use and Interpretation. Anesth Analg. 2018;126:1763–8.
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, et al. Cell Miner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009;10:277.
Luchini C, Bibeau F, Ligtenberg MJL, Singh N, Nottegar A, Bosse T, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol. 2019;30:1232–43.
Rizzo A, Ricci AD, Brandi G. PD-L1, TMB, MSI, and other predictors of response to immune checkpoint inhibitors in biliary tract cancer. Cancers. 2021;13:558.
Condelli V, Calice G, Cassano A, Basso M, Rodriquenz MG, Zupa A, et al. Novel epigenetic eight-gene signature predictive of poor prognosis and MSI-like phenotype in human metastatic colorectal carcinomas. Cancers. 2021;13:158.
Morreale FE, Walden H. Types of ubiquitin ligases. Cell. 2016;165:248-e1.
Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Res. 2016;26:423–40.
Chen L, Min J, Wang F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct Target Ther. 2022;7:378.
Tong X, Tang R, Xiao M, Xu J, Wang W, Zhang B, et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. 2022;15:174.
Li J, Wu F, Li C, Sun S, Feng C, Wu H, et al. The cuproptosis-related signature predicts prognosis and indicates immune microenvironment in breast cancer. Front Genet. 2022;13: 977322.
Winkler GS, Albert TK, Dominguez C, Legtenberg YI, Boelens R, Timmers HT. An altered-specificity ubiquitin-conjugating enzyme/ubiquitin-protein ligase pair. J Mol Biol. 2004;337:157–65.
Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, et al. Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IkappaBalpha. J Biol Chem. 1999;274:14823–30.
Chiang MH, Chen LF, Chen H. Ubiquitin-conjugating enzyme UBE2D2 is responsible for FBXW2 (F-box and WD repeat domain containing 2)-mediated human GCM1 (glial cell missing homolog 1) ubiquitination and degradation. Biol Reprod. 2008;79:914–20.
Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S. The N terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin-conjugating enzyme. J Biol Chem. 2010;285:23687–98.
Smith J, Sen S, Weeks RJ, Eccles MR, Chatterjee A. Promoter DNA hypermethylation and paradoxical gene activation. Trends Cancer. 2020;6:392–406.
Suski JM, Braun M, Strmiska V, Sicinski P. Targeting cell-cycle machinery in cancer. Cancer Cell. 2021;39:759–78.
Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12:71.
Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308.
Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol. 2018;13:395–412.
Monteith GR, Prevarskaya N, Roberts-Thomson SJ. The calcium-cancer signalling nexus. Nat Rev Cancer. 2017;17:367–80.
Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7.
Liu J, Peng Y, Wei W. Cell cycle on the crossroad of tumorigenesis and cancer therapy. Trends Cell Biol. 2022;32:30–44.
Kovalski JR, Kuzuoglu-Ozturk D, Ruggero D. Protein synthesis control in cancer: selectivity and therapeutic targeting. EMBO J. 2022;41: e109823.
Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24:2482–90.
Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84.
Du W, Frankel TL, Green M, Zou W. IFNgamma signaling integrity in colorectal cancer immunity and immunotherapy. Cell Mol Immunol. 2022;19:23–32.
Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 2020;10:26–39.
Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer. 2021;21:298–312.
Xun Y, Yang H, Kaminska B, You H. Toll-like receptors and toll-like receptor-targeted immunotherapy against glioma. J Hematol Oncol. 2021;14:176.
Jiang Y, Zhang H, Wang J, Chen J, Guo Z, Liu Y, et al. Exploiting RIG-I-like receptor pathway for cancer immunotherapy. J Hematol Oncol. 2023;16:8.
Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(548–63): e16.
Zhang F, Wang H, Yu J, Yao X, Yang S, Li W, et al. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer. 2021;20:6.
He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020;30:660–9.
Propper DJ, Balkwill FR. Harnessing cytokines and chemokines for cancer therapy. Nat Rev Clin Oncol. 2022;19:237–53.
Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–33.
Lee YS, Radford KJ. The role of dendritic cells in cancer. Int Rev Cell Mol Biol. 2019;348:123–78.
Zhang Y, Zou J, Chen R. An M0 macrophage-related prognostic model for hepatocellular carcinoma. BMC Cancer. 2022;22:791.
Yoshitomi H, Ueno H. Shared and distinct roles of T peripheral helper and T follicular helper cells in human diseases. Cell Mol Immunol. 2021;18:523–7.
Wang Y, Wang C, Qiu J, Qu X, Peng J, Lu C, et al. Targeting CD96 overcomes PD-1 blockade resistance by enhancing CD8+ TIL function in cervical cancer. J Immunother Cancer. 2022;10:e003667.
Li H, Wu X, Cheng X. Advances in diagnosis and treatment of metastatic cervical cancer. J Gynecol Oncol. 2016;27:e43.
Mendhiratta N, Muraki P, Sisk AE Jr, Shuch B. Papillary renal cell carcinoma: review. Urol Oncol. 2021;39:327–37.
Funding
This work was supported by the National Natural Science Foundation of China (82373155, 82103293, 82172651), the Natural Science Foundation of Anhui Province (2108085MH287), the Natural Science Foundation of Anhui Education Department for Distinguished Young Scholars (2022AH020074), the Natural Science Foundation of Anhui Education Department for Excellent Young Scholars (2022AH030123), the Support Plan for Outstanding Young Talents of Anhui Education Department (gxyq2021257), the Key Research and Development Project of Anhui Province (202104j07020019), the Science and Technology Project of Wuhu City (2022jc52) and the Talent Introduction Science Foundation of Yijishan Hospital, Wannan Medical College (No. YR202109 and YR202110).
Author information
Authors and Affiliations
Contributions
YF: conceptualization, data curation, formal analysis, investigation, methodology, software, validation, writing—original draft preparation, writing—review and editing. DC: conceptualization, data curation, formal analysis, methodology, validation, visualization, writing—original draft preparation, writing—review and editing. RD: methodology, software, validation. YL: data curation, investigation, supervision, validation. ZW: investigation, supervision, visualization. PG: software, supervision, visualization. MZ: software, supervision, visualization. XW: supervision, writing—review and editing. XZ: conceptualization, data curation, methodology, resources, validation, writing—review and editing, funding acquisition. JC: conceptualization, project administration, resources, validation, funding acquisition.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Ethical approval
The manuscript does not contain clinical studies or patient data.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fei, Y., Cao, D., Dong, R. et al. The cuproptosis-related gene UBE2D2 functions as an immunotherapeutic and prognostic biomarker in pan-cancer. Clin Transl Oncol (2024). https://doi.org/10.1007/s12094-024-03495-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12094-024-03495-4