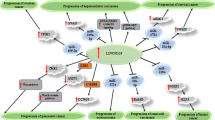
Abstract
Long non-coding RNAs (lncRNAs) are involved the progression of cancerous and non-cancerous disorders via different mechanism. FTX (five prime to xist) is an evolutionarily conserved lncRNA that is located upstream of XIST and regulates its expression. FTX participates in progression of various malignancy including gastric cancer, glioma, ovarian cancer, pancreatic cancer, and retinoblastoma. Also, FTX can be involved in the pathogenesis of non-cancerous disorders such as endometriosis and stroke. FTX acts as competitive endogenous RNA (ceRNA) and via sponging various miRNAs, including miR-186, miR-200a-3p, miR-215-3p, and miR-153-3p to regulate the expression of their downstream target. FTX by targeting various signaling pathways including Wnt/β-catenin, PI3K/Akt, SOX4, PDK1/PKB/GSK-3β, TGF-β1, FOXA2, and PPARγ regulate molecular mechanism involved in various disorders. Dysregulation of FTX is associated with an increased risk of various disorders. Therefore, FTX and its downstream targets may be suitable biomarkers for the diagnosis and treatment of human malignancies. In this review, we summarized the emerging roles of FTX in human cancerous and non-cancerous cells.






Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Gu L, Li Q, Liu H, Lu X, Zhu M. Long noncoding RNA TUG1 promotes autophagy-associated paclitaxel resistance by sponging miR-29b-3p in ovarian cancer cells. Onco Targets Ther. 2007;2020:13.
Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354–66.
Farzaneh M, Ghasemian M, Ghaedrahmati F, Poodineh J, Najafi S, Masoodi T, et al. Functional roles of lncRN-TUG1 in hepatocellular carcinoma. Life Sci. 2022;308:120974.
Xu W-W, Jin J, Wu X-y, Ren Q-L, Farzaneh M. MALAT1-related signaling pathways in colorectal cancer. Cancer Cell Int. 2022;22:1–9.
Ghasemian M, Rajabibazl M, Sahebi U, Sadeghi S, Maleki R, Hashemnia V, et al. Long non-coding RNA MIR4435-2HG: a key molecule in progression of cancer and non-cancerous disorders. Cancer Cell Int. 2022;22:1–12.
Jing D, Zhu F, Xu Z, Zhang G, Zhou G. The role of long noncoding RNA (lncRNA) nuclear-enriched abundant transcript 1 (NEAT1) in immune diseases. Transpl Immunol. 2022;75:101716.
Vimalraj S, Subramanian R. Biogenesis, classification, and role of LncRNAs in tumor angiogenesis: a focus on tumor and its neighbouring cells, and interaction with miRNAs. Process Biochem. 2022;122:347.
Alsaedy HK, Mirzaei AR, Alhashimi RAH. Investigating the structure and function of Long Non-Coding RNA (LncRNA) and its role in cancer. Cell Mol Biomed Rep. 2022;2:245–53.
Ghasemian M, Rajabibazl M, Mirfakhraie R, Razavi AE, Sadeghi H. Long noncoding RNA LINC00978 acts as a potential diagnostic biomarker in patients with colorectal cancer. Exp Mol Pathol. 2021;122: 104666.
Azizidoost S, Ghaedrahmati F, Anbiyaee O, Ahmad Ali R, Cheraghzadeh M, Farzaneh M. Emerging roles for lncRNA-NEAT1 in colorectal cancer. Cancer Cell Int. 2022;22:1–10.
Marx V. How noncoding RNAs began to leave the junkyard. Nat Methods. 2022;10:1–4.
Haridevamuthu B, Guru A, Velayutham M, Snega Priya P, Arshad A, Arockiaraj J. Long non-coding RNA, a supreme post-transcriptional immune regulator of bacterial or virus-driven immune evolution in teleost. Rev Aquac. 2022;15:163.
Zhang Q, Zhong C, Shen J, Chen S, Jia Y, Duan S. Emerging role of LINC00461 in cancer. Biomed Pharmacother. 2022;152: 113239.
Xie Z, Zhong C, Shen J, Jia Y, Duan S. LINC00963: a potential cancer diagnostic and therapeutic target. Biomed Pharmacother. 2022;150: 113019.
Ratti M, Lampis A, Ghidini M, Salati M, Mirchev MB, Valeri N, et al. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Target Oncol. 2020;15:261–78.
Connerty P, Lock RB, De Bock CE. Long non-coding RNAs: major regulators of cell stress in cancer. Front Oncol. 2020;10:285.
Jin K-T, Yao J-Y, Fang X-L, Di H, Ma Y-Y. Roles of lncRNAs in cancer: focusing on angiogenesis. Life Sci. 2020;252: 117647.
Sahin Y. LncRNA H19 is a potential biomarker and correlated with immune infiltration in thyroid carcinoma. Clin Exp Med. 2022. https://doi.org/10.1007/s10238-022-00853-w.
Lu L, Huang J, Mo J, Da X, Li Q, Fan M, et al. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol Biol Lett. 2022;27:17.
Altan Z, Sahin Y. miR-203 suppresses pancreatic cancer cell proliferation and migration by modulating DUSP5 expression. Mol Cell Probes. 2022;66: 101866.
Wang J, Mo J, Xie Y, Wang C. Ultrasound microbubbles-mediated miR-216b affects MALAT1-miRNA axis in non-small cell lung cancer cells. Tissue Cell. 2022;74: 101703.
Santos-Rebouças CB, Boy R, Vianna EQ, Gonçalves AP, Piergiorge RM, Abdala BB, et al. Skewed X-chromosome inactivation and compensatory upregulation of escape genes precludes major clinical symptoms in a female with a large Xq deletion. Front Genet. 2020;11:101.
Hierholzer A, Chureau C, Liverziani A, Ruiz NB, Cattanach BM, Young AN, et al. A long noncoding RNA influences the choice of the X chromosome to be inactivated. Proc Natl Acad Sci. 2022;119: e2118182119.
Siniscalchi C, Di Palo A, Russo A, Potenza N. The lncRNAs at X chromosome inactivation center: not just a matter of sex dosage compensation. Int J Mol Sci. 2022;23:611.
Furlan G, Hernandez NG, Huret C, Galupa R, van Bemmel JG, Romito A, et al. The Ftx noncoding locus controls X chromosome inactivation independently of its RNA products. Mol Cell. 2018;70(462–472): e468.
Liu F, Yuan J, Huang J, Yang F, Wang T, Ma J, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–34.
Furlan G, Gutierrez Hernandez N, Huret C, Galupa R, van Bemmel JG, Romito A, et al. The Ftx noncoding locus controls X chromosome inactivation independently of its RNA products. Mol Cell. 2018;70:462-472.e468.
Chanda K, Mukhopadhyay D. LncRNA Xist, X-chromosome instability and Alzheimer’s disease. Curr Alzheimer Res. 2020;17:499–507.
Santos-Rebouças CB. Epigenetics of X-chromosome Inactivation. In: Handbook of Epigenetics. Elsevier: Amsterdam; 2023. p. 419–41.
Yang J, Qu T, Li Y, Ma J, Yu H. Biological role of long non-coding RNA FTX in cancer progression. Biomed Pharmacother. 2022;153: 113446.
Zhao Q, Li T, Qi J, Liu J, Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS ONE. 2014;9: e109782.
Wang X, Su Y, Yin C. Long non-coding RNA (lncRNA) five prime to Xist (FTX) promotes retinoblastoma progression by regulating the microRNA-320a/with-no-lysine kinases 1 (WNK1) axis. Bioengineered. 2021;12:11622–33.
Zhang Y, Fan X, Yang H. Long noncoding RNA FTX ameliorates hydrogen peroxide-induced cardiomyocyte injury by regulating the miR-150/KLF13 axis. Open Life Sci. 2020;15:1000–12.
Pan L, Du M, Liu H, Cheng B, Zhu M, Jia B, et al. LncRNA FTX promotes the malignant progression of colorectal cancer by regulating the miR-214–5p–JAG1 axis. Ann Transl Med. 2021;9:1369.
Zhao K, Ye Z, Li Y, Li C, Yang X, Chen Q, et al. LncRNA FTX contributes to the progression of colorectal cancer through regulating miR-192-5p/EIF5A2 axis. Onco Targets Ther. 2020;13:2677.
Yang Y, Zhang J, Chen X, Xu X, Cao G, Li H, et al. LncRNA FTX sponges miR-215 and inhibits phosphorylation of vimentin for promoting colorectal cancer progression. Gene Ther. 2018;25:321–30.
Zhang F, Wang X, Tang B, Li P, Wen Y, Yu P. Long non-coding RNA FTX promotes gastric cancer progression by targeting miR-215. Eur Rev Med Pharmacol Sci. 2020;24:3037–48.
Liu L, Li X, Shi Y, Chen H. The long noncoding RNA FTX promotes a malignant phenotype in bone marrow mesenchymal stem cells via the miR-186/c-Met axis. Biomed Pharmacother. 2020;131: 110666.
Liang Y, Lu H. Long noncoding RNA FTX is associated with prognosis of glioma patients. J Gene Med. 2020;22: e3237.
Chen Z, Zhang M, Lu Y, Ding T, Liu Z, Liu Y, et al. Overexpressed lncRNA FTX promotes the cell viability, proliferation, migration and invasion of renal cell carcinoma via FTX/miR-4429/UBE2C axis. Oncol Rep. 2022;48:1–15.
Liu M, Peng J. FTX regulated miR-153–3p/FOXR2 to promote cisplatin resistance in ovarian cancer. Comput Math Methods Med. 2022;2022:1.
Li S, Zhang Q, Liu W, Zhao C. Silencing of FTX suppresses pancreatic cancer cell proliferation and invasion by upregulating miR-513b-5p. BMC Cancer. 2021;21:1–12.
Chen H, Liu T, Ouyang H, Lin S, Zhong H, Zhang H, et al. Upregulation of FTX promotes osteosarcoma tumorigenesis by increasing SOX4 expression via miR-214-5p. Onco Targets Ther. 2020;13:7125.
Yang X, Tao H, Wang C, Chen W, Hua F, Qian H. lncRNA-ATB promotes stemness maintenance in colorectal cancer by regulating transcriptional activity of the β-catenin pathway. Exp Ther Med. 2020;19:3097–103.
Gao Q, Wang Y. LncRNA FTX regulates angiogenesis through miR-342-3p/SPI1 axis in stroke. Neuropsychiatr Dis Treat. 2021;17:3617.
Hosoi Y, Soma M, Shiura H, Sado T, Hasuwa H, Abe K, et al. Female mice lacking Ftx lncRNA exhibit impaired X-chromosome inactivation and a microphthalmia-like phenotype. Nat Commun. 2018;9:1–13.
Long B, Li N, Xu X-X, Li X-X, Xu X-J, Guo D, et al. Long noncoding RNA FTX regulates cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2l2. Biochem Biophys Res Commun. 2018;495:312–8.
Poodineh J, Sirati-Sabet M, Rajabibazl M, Ghasemian M, Mohammadi-Yeganeh S. Downregulation of NRARP exerts anti-tumor activities in the breast tumor cells depending on Wnt/β-catenin-mediated signals: The role of miR-130a-3p. Chem Biol Drug Des. 2022;100:334–45.
Jin S, He J, Zhou Y, Wu D, Li J, Gao W. LncRNA FTX activates FOXA2 expression to inhibit non–small-cell lung cancer proliferation and metastasis. J Cell Mol Med. 2020;24:4839–49.
Shen Y, Yang G, Zhuo S, Zhuang H, Chen S. lncRNA FTX promotes asthma progression by sponging miR-590-5p and upregulating JAK2. Am J Transl Res. 2021;13:8833.
Yang X, Tao L, Zhu J, Zhang S. Long noncoding RNA FTX reduces hypertrophy of neonatal mouse cardiac myocytes and regulates the PTEN/PI3K/Akt signaling pathway by sponging microRNA-22. Med Sci Monit. 2019;25:9609.
Zuo Y, Sun H, Song L, Hu Y, Guo F. LncRNA FTX involves in the Nogo-66-induced inhibition of neurite outgrowth through regulating PDK1/PKB/GSK-3β pathway. Cell Mol Neurobiol. 2020;40:1143–53.
Li X, Zhao Q, Qi J, Wang W, Zhang D, Li Z, et al. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARγ pathway in hepatocellular carcinoma. Int J Oncol. 2018;53:551–66.
Choudhary S, Burns SC, Mirsafian H, Li W, Vo DT, Qiao M, et al. Genomic analyses of early responses to radiation in glioblastoma reveal new alterations at transcription, splicing, and translation levels. Sci Rep. 2020;10:1–12.
He X, Sun F, Guo F, Wang K, Gao Y, Feng Y, et al. Knockdown of long noncoding RNA FTX inhibits proliferation, migration, and invasion in renal cell carcinoma cells. Oncol Res. 2017;25:157.
Huang S, Zhu X, Ke Y, Xiao D, Liang C, Chen J, et al. LncRNA FTX inhibition restrains osteosarcoma proliferation and migration via modulating miR-320a/TXNRD1. Cancer Biol Ther. 2020;21:379–87.
Li B, Ren P, Wang Z. Long non-coding RNA Ftx promotes osteosarcoma progression via the epithelial to mesenchymal transition mechanism and is associated with poor prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2018;11:4503.
Yu Y, Yao P, Wang Z, Xie W. Down-regulation of FTX promotes the differentiation of osteoclasts in osteoporosis through the Notch1 signaling pathway by targeting miR-137. BMC Musculoskelet Disord. 2020;21:1–10.
Wang P, Zhang Y, Wang W, Jiang H. Upregulation of FTX expression is associated with a poor prognosis and contributes to the progression of thyroid cancer. Oncol Lett. 2021;22:1–9.
Luzón-Toro B, Villalba-Benito L, Fernández RM, Torroglosa A, Antiñolo G, Borrego S. RMRP, RMST, FTX and IPW: novel potential long non-coding RNAs in medullary thyroid cancer. Orphanet J Rare Dis. 2021;16:1–7.
Huo X, Wang H, Huo B, Wang L, Yang K, Wang J, et al. FTX contributes to cell proliferation and migration in lung adenocarcinoma via targeting miR-335-5p/NUCB2 axis. Cancer Cell Int. 2020;20:1–13.
Zhang W, Bi Y, Li J, Peng F, Li H, Li C, et al. Long noncoding RNA FTX is upregulated in gliomas and promotes proliferation and invasion of glioma cells by negatively regulating miR-342-3p. Lab Invest. 2017;97:447–57.
Li H, Yao G, Zhai J, Hu D, Fan Y. LncRNA FTX promotes proliferation and invasion of gastric cancer via miR-144/ZFX axis. Onco Targets Ther. 2019;12:11701.
Zhu L, Jia R, Zhang J, Li X, Qin C, Zhao Q. Quantitative proteomics analysis revealed the potential role of lncRNA ftx in promoting gastric cancer progression. Proteom Clin Appl. 2020;14:1900053.
Guo X-B, Hua Z, Li C, Peng L-P, Wang J-S, Wang B, et al. Biological significance of long non-coding RNA FTX expression in human colorectal cancer. Int J Clin Exp Med. 2015;8:15591.
Chen G-Q, Liao Z-M, Liu J, Li F, Huang D, Zhou Y-D. LncRNA FTX promotes colorectal cancer cells migration and invasion by miRNA-590-5p/RBPJ axis. Biochem Genet. 2021;59:560–73.
Jia R, Song L, Fei Z, Qin C, Zhao Q. Long noncoding RNA Ftx regulates the protein expression profile in HCT116 human colon cancer cells. Proteome Sci. 2022;20:1–12.
Kwok ZH, Zhang B, Chew XH, Chan JJ, Teh V, Yang H, et al. Systematic analysis of intronic miRNAs reveals cooperativity within the multicomponent FTX locus to promote colon cancer development. Can Res. 2021;81:1308–20.
Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350.
Wu H, Zhong Z, Wang A, Yuan C, Ning K, Hu H, et al. LncRNA FTX represses the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via regulating the M1/M2 polarization of Kupffer cells. Cancer Cell Int. 2020;20:1–11.
Wang H, Ni C, Xiao W, Wang S. Role of lncRNA FTX in invasion, metastasis, and epithelial-mesenchymal transition of endometrial stromal cells caused by endometriosis by regulating the PI3K/Akt signaling pathway. Ann Transl Med. 2020;8:1504.
Aegerter H, Lambrecht BN. The pathology of asthma: what is obstructing our view? Annu Rev Pathol. 2023;18:387–409.
Hong H, Li K, Wei X: OCT4A and its related LncRNA FTX modulate the self-renewal of dental pulp cells under inflammatory microenvironment. In 第十一届全国化学生物学学术会议论文摘要 (第二卷). 2019
Xiang W, Jiang L, Zhou Y, Li Z, Zhao Q, Wu T, et al. The lncRNA Ftx/miR-382-5p/Nrg1 axis improves the inflammation response of microglia and spinal cord injury repair. Neurochem Int. 2021;143: 104929.
Liu X, Li C, Zhu J, Li W, Zhu Q. Dysregulation of FTX/miR-545 signaling pathway downregulates Tim-3 and is responsible for the abnormal activation of macrophage in cirrhosis. J Cell Biochem. 2019;120:2336–46.
Feigin VL, Brainin M, Norrving B, Martins S, Sacco RL, Hacke W, et al. World stroke organization (WSO): global stroke fact sheet 2022. Int J Stroke. 2022;17:18–29.
Gao Q, Wang Y. LncRNA FTX regulates angiogenesis through miR-342–3p/SPI1 Axis in stroke. Neuropsychiatr Dis Treat. 2021;17:3617–25.
Sun X, Jiang Y, Li Q, Tan Q, Dong M, Cai Be, et al. Quantitative proteomics analysis revealed the potential role of lncRNA Ftx in cardiomyocytes. Proteome Sci. 2023;21:2.
Yang X, Tao L, Zhu J, Zhang S. Long noncoding RNA FTX reduces hypertrophy of neonatal mouse cardiac myocytes and regulates the PTEN/PI3K/Akt signaling pathway by sponging microRNA-22. Med Sci Monit. 2019;25:9609–17.
Li L, Li L, Zhang Y, Yang H, Wang Y. Long non-coding RNA FTX alleviates hypoxia/reoxygenation-induced cardiomyocyte injury via miR-410-3p/Fmr1 axis. Eur Rev Med Pharmacol Sci. 2020;24:396–408.
Cao P, Zhao B, Xiao Y, Hu S, Kong K, Han P, et al. Understanding the critical role of glycolysis-related lncRNAs in lung adenocarcinoma based on three molecular subtypes. BioMed Research International. 2022;2022:1.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors have approved the submitted version of the article and have agreed to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Informed consent
As this paper is just a review paper, the informed consent is not necessary.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sheykhi-Sabzehpoush, M., Ghasemian, M., Khojasteh Pour, F. et al. Emerging roles of long non-coding RNA FTX in human disorders. Clin Transl Oncol 25, 2812–2831 (2023). https://doi.org/10.1007/s12094-023-03163-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-023-03163-z