Abstract
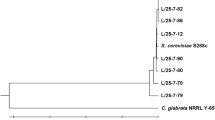
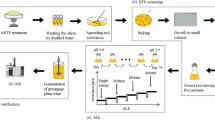
A wild-type strain was isolated from slightly rotted pears after three rounds of enrichment culture, identified as Saccharomyces cerevisiae 3308, and evaluated for its fermentation capability of second generation bioethanol and tolerance of temperature, glucose and ethanol. S. cerevisiae 3308 was mutated by using the physical and chemical mutagenesis methods, ultraviolet (UV) and diethyl sulfate (DES), respectively. Positive mutated strains were mainly generated by the treatment of UV, but numerous negative mutations emerged under the treatment of DES. A positive mutated strain, UV-20, produced ethanol from 62.33 ± 1.34 to 122.22 ± 2.80 g/L at 30–45 °C, and had a maximum yield of ethanol at 37 °C. Furthermore, UV-20 produced 121.18 ± 2.51 g/L of second generation bioethanol at 37 °C. Simultaneously, UV-20 exhibited superior tolerance to 50% of glucose and 21% of ethanol. In a conclusion, all of these results indicated that UV-20 has a potential industrial application value.







Similar content being viewed by others
References
Dasgupta D, Ghosh D, Bandhu S et al (2017) Lignocellulosic sugar management for xylitol and ethanol fermentation with multiple cell recycling by Kluyveromyces marxianus IIPE453. Microbiol Res 200:64–72. https://doi.org/10.1016/j.micres.2017.04.002
Díaz-Nava LE, Montes-Garcia N, Domínguez JM et al (2017) Effect of carbon sources on the growth and ethanol production of native yeast Pichia kudriavzevii ITV-S42 isolated from sweet sorghum juice. Bioproc Biosyst Eng. https://doi.org/10.1007/s00449-017-1769-z
Arshad M, Ahmed S, Zia MA et al (2014) Kinetics and thermodynamics of ethanol production by Saccharomyces cerevisiae MLD10 using molasses. Appl Biochem Biotechnol 172:2455–2464. https://doi.org/10.1007/s12010-013-0689-x
Henderson CM, Block DE (2014) Examining the role of membrane lipid composition in determining the ethanol tolerance of Saccharomyces cerevisiae. Appl Environ Microb 80:2966–2972. https://doi.org/10.1128/AEM.04151-13
Snoek T, Verstrepen KJ, Voordeckers K (2016) How do yeast cells become tolerant to high ethanol concentrations? Curr Genet 62:475–480. https://doi.org/10.1007/s00294-015-0561-3
Dhaliwal SS, Oberoi HS, Sandhu SK et al (2011) Enhanced ethanol production from sugarcane juice by galactose adaptation of a newly isolated thermotolerant strain of Pichiakudriavzevii. Bioresour Technol 102:5968–5975. https://doi.org/10.1016/j.biortech.2011.02.015
Oberoi HS, Babbar N, Sandhu SK et al (2012) Ethanol production from alkali-treated rice straw via simultaneous saccharification and fermentation using newly isolated thermotolerant Pichia kudriavzevii HOP-1. J Ind Microbiol Biotechnol 39:557–566. https://doi.org/10.1007/s10295-011-1060-2
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998 73:331–371. https://doi.org/10.1023/A:1001761008817
Gu H, Zhang J, Bao J (2014) Inhibitor analysis and adaptive evolution of Saccharomyces cerevisiae for simultaneous saccharification and ethanol fermentation from industrial waste corncob residues. Bioresour Technol 157:6–13. https://doi.org/10.1016/j.biortech.2014.01.060
Cheng CL, Chang JS (2011) Hydrolysis of lignocellulosic feedstock by novel cellulases originating from Pseudomonas sp. CL3 for fermentative hydrogen production. Bioresour Technol 102:8628–8634. https://doi.org/10.1016/j.biortech.2011.03.053
Kitichantaropas Y, Boonchird C, Sugiyama M et al (2016) Cellular mechanisms contributing to multiple stress tolerance in Saccharomyces cerevisiae strains with potential use in high-temperature ethanol fermentation. AMB Express 6:107. https://doi.org/10.1186/s13568-016-0285-x
Zhao X, Wang W, Tong B, Zhang S, Wei D (2016) A newly isolated Penicillium oxalicum 16 cellulase with high efficient synergism and high tolerance of Monosaccharide. Appl Biochem Biotechnol 178:173–183. https://doi.org/10.1007/s12010-015-1866-x
Roy U, Jessani LG, Rudramurthy SM et al (2017) Seven cases of Saccharomyces fungaemia related to use of probiotics. Mycoses 60:375–380. https://doi.org/10.1111/myc.12604
Alexandre H (2013) Flor yeasts of Saccharomyces cerevisiae—their ecology, genetics and metabolism. Int J Food Microbiol 167:269–275. https://doi.org/10.1016/j.ijfoodmicro.2013.08.021
Ren G, Ma A, Liu W et al (2016) Bacterial signals N-acyl homoserine lactones induce the changes of morphology and ethanol tolerance in Saccharomyces cerevisiae. AMB Express 6:117. https://doi.org/10.1186/s13568-016-0292-y
Baselga I, Zafra O, Lago EP et al (2017) An AFLP based method for the detection and identification of indigenous yeast in complex must samples without a microbiological culture. Int J Food Microbiol 241:89–97. https://doi.org/10.1016/j.ijfoodmicro.2016.09.014
Li X, Liu R, Li J et al (2015) Enhanced arachidonic acid production from Mortierella alpina combining atmospheric and room temperature plasma (ARTP) and diethyl sulfate treatments. Bioresour Technol 177:134–140. https://doi.org/10.1016/j.biortech.2014.11.051
Wu CJ, Li CW, Cui CB (2014) Seven new and two known lipopeptides as well as five known polyketides: the activated production of silent metabolites in a marine-derived fungus by chemical mutagenesis strategy using diethyl sulphate. Mar Drugs 12:1815–1838. https://doi.org/10.3390/md12041815
Watanabe T, Watanabe I, Yamamoto M et al (2011) A UV-induced mutant of Pichia stipitis with increased ethanol production from xylose and selection of a spontaneous mutant with increased ethanol tolerance. Bioresour Technol 102:1844–1848. https://doi.org/10.1016/j.biortech.2010.09.087
Woo JM, Yang KM, Kim SU et al (2014) High temperature stimulates acetic acid accumulation and enhances the growth inhibition and ethanol production by Saccharomyces cerevisiae under fermenting conditions. Appl Microbiol Biotechnol 98:6085–6094. https://doi.org/10.1007/s00253-014-5691-x
Singh A, Bajar S, Bishnoi NR (2014) Enzymatic hydrolysis of microwave alkali pretreated rice husk for ethanol production by Saccharomyces cerevisiae, Scheffersomyces stipitis and their co-culture. Fuel 116:699–702. https://doi.org/10.1016/j.fuel.2013.08.072
Huang Y, Qin X, Luo XM et al (2015) Efficient enzymatic hydrolysis and simultaneous saccharification and fermentation of sugarcane bagasse pulp for ethanol production by cellulase from Penicillium oxalicum EU2106 and thermotolerant Saccharomyces cerevisiae ZM1-5. Biomass Bioenergy 77:53–63. https://doi.org/10.1016/j.biombioe.2015.03.020
Snoek T, Nicolino MP, Van den Bremt S et al (2015) Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance. Biotechnol Biofuels 8:32. https://doi.org/10.1186/s13068-015-0216-0
Yusuf F, Gaur NA (2016) Metabolic and adaptive laboratory evolutionary engineering (ALE) of Saccharomyces cerevisiae for second generation biofuel production. IJEPE. https://doi.org/10.1999/1307-6892/60367
Lu Y, Voon MKW, Huang D et al (2017) Combined effects of fermentation temperature and pH on kinetic changes of chemical constituents of durian wine fermented with Saccharomyces cerevisiae. Appl Microbiol Biotechnol 101:3005–3014. https://doi.org/10.1007/s00253-016-8043-1
Zhao F, Du Y, Bai P et al (2017) Enhancing Saccharomyces cerevisiae reactive oxygen species and ethanol stress tolerance for high-level production of protopanoxadiol. Bioresour Technol 227:308–316. https://doi.org/10.1016/j.biortech.2016.12.061
Dalawai N, Krupa KN, Nadkarni S et al (2017) Screening of efficient ethanol tolerant yeast strain for production of ethanol. Int J Pure Appl Biosci 5:744–752. https://doi.org/10.18782/2320-7051.2587
Qiu Z, Jiang R (2017) Improving Saccharomyces cerevisiae ethanol production and tolerance via RNA polymerase II subunit Rpb7. Biotechnol Biofuels 10:125. https://doi.org/10.1186/s13068-017-0806-0
Ikehata H, Ono T (2011) The mechanisms of UV mutagenesis. J Radiat Res 52:115–125. https://doi.org/10.1269/jrr.10175
Landaud S, Helinck S, Bonnarme P (2008) Formation of volatile sulfur compounds and metabolism of methionine and other sulfur compounds in fermented food. Appl Microbiol Biotechnol 77:1191–1205. https://doi.org/10.1007/s00253-007-1288-y
Torija MJ, Rozes N, Poblet M et al (2003) Effects of fermentation temperature on the strain population of Saccharomyces cerevisiae. Int J Food Microbiol 80:47–53. https://doi.org/10.1016/S0168-1605(02)00144-7
Acknowledgements
This work was funded by National Natural Science Foundation of China (21666010 and 31360217), and Doctoral Starting up Foundation of Jiangxi Normal University (5451).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yi, S., Zhang, X., Li, Hx. et al. Screening and Mutation of Saccharomyces cerevisiae UV-20 with a High Yield of Second Generation Bioethanol and High Tolerance of Temperature, Glucose and Ethanol. Indian J Microbiol 58, 440–447 (2018). https://doi.org/10.1007/s12088-018-0741-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-018-0741-1