Abstract
Background
Simeprevir with peginterferon and ribavirin has been used for the treatment of chronic hepatitis caused by genotype 1 hepatitis C virus (HCV). We explored the predictive factors for sustained virological response (SVR) and viral relapse using datasets from four Japanese phase 3 studies (CONCERTO).
Methods
We used a multiple logistic regression model. First, an integrated dataset comprising 357 patients was analyzed. Subsequently, prior treatment-naïve and relapser (223 patients) and nonresponder (134 patients) of interferon-based treatment subsets were analyzed to identify predictors of SVR. A subset of nonresponders (106 patients) who were treated ≥24 weeks was also analyzed to identify predictors for viral relapse.
Results
In the integrated dataset, prior treatment response was significantly associated with SVR. In subset analyses, interleukin-28B (IL28B) TT genotype and undetectable plasma HCV RNA level at week 4 were associated in treatment-naïve patients and relapsers [odds ratio (OR); 4.106 and 3.701, respectively]. In the nonresponders, the IL28B TT genotype population was very small, and inosine triphosphatase (ITPA) and undetectable plasma HCV RNA at week 4 were associated (OR; 2.506 and 3.333, respectively). Furthermore, ribavirin dose intensity (RBV-DI) and detectable plasma HCV RNA at week 4 were significantly associated with viral relapse (OR; 0.327 and 2.922, respectively).
Conclusion
IL28B and plasma HCV RNA level at week 4 were clinically relevant predictive factors for SVR in treatment-naïve patients and relapsers. Moreover, RBV-DI and plasma HCV level at week 4 were identified as relevant predictive factors for viral relapse in nonresponders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis C virus (HCV) infection is one of the major concerns for public health, with approximately >180 million infections worldwide [1]. During the natural course of HCV infection, patients develop liver fibrosis, which gradually progresses to hepatocellular carcinoma in approximately 45 % of cases within 20 years from the initial infection [2]. Because interferons (IFNs) have been known to have potential eradication of the HCV, they have been used as the standard of care for HCV treatment [3, 4].
Sustained virological response (SVR), defined as undetectable HCV RNA by quantification assay at the end of treatment and 12–24 weeks after the end of treatment, has been used to assess therapeutic efficacy. In global HCV treatment guidelines, SVR is recommended as the goal of anti-HCV treatment, patients who achieved SVR experienced numerous benefits including a reduction of liver fibrosis and hepatocellular carcinoma [5, 6].
Over the past decade, the combination of peginterferon (PegIFN) plus ribavirin (RBV), developed for genotype 1 (GT1) HCV-infected patients, has conferred SVR rate of approximately 40–50 % for treatment-naïve GT1 HCV patients with high viral load [7, 8]. The limited SVR rates observed with this drug combination reiterated the importance of predictive factors for SVR when identifying appropriate patients that would benefit from this combination treatment, thereby necessitating the development of a new active agent [9–11]. Telaprevir is the first-generation oral NS3/4A protease inhibitor which was developed in combination with PegIFN plus RBV for its superior treatment efficacy. The SVR rates at 24 weeks after the end of treatment (SVR24) in treatment-naïve as well as prior IFN-based therapy relapser and nonresponder GT1 HCV patients were approximately 75, 83, and 41 %, respectively [12, 13].
Simeprevir [100 mg once daily (QD)] is a second-generation oral NS3/4A protease inhibitor, which was developed in combination with PegIFN and RBV. Four phase 3 studies (CONCERTO) were conducted in Japan. The SVR24 rates were 88.6 and 91.7 % for treatment-naïve; 35.8, 50.9, and 38.5 % for nonresponders; and 89.8 and 96.6 % for relapser patients [14–16]. Because simeprevir has favorable efficacy without inducing severe dermatologic and hematologic toxicity, it provides a better therapeutic index in GT1 HCV patients. Unfortunately, certain treatment failures have been reported: SVR24 was not favorable in nonresponder patients even after the achievement of an early viral response, and approximately 40 % of patients exhibited relapse after discontinuation of therapy [15]. The present study aimed to identify potential predictive factors for SVR24 and viral relapse to determine the patients most suitable to receive the combination of simeprevir with PegIFN and RBV by using a consolidated data set from the aforementioned CONCERTO studies.
Methods
Data source
The data for this post hoc study was derived from four phase 3 studies to assess the efficacy and safety of simeprevir plus PegIFN and RBV for the treatment of GT1 HCV-infected patients (CONCERTO-1, NCT01292239; CONCERTO-2, NCT01288209; CONCERTO-3, NCT01290731; and CONCERTO-4, NCT01366638). All patients who received simeprevir were included in this study. Original results from each CONCERTO study are available in the literature [14–16].
The CONCERTO studies included the following target population: CONCERTO-1, treatment-naïve patients (naïve); CONCERTO-2, prior nonresponders to IFN-based therapy (nonresponders); CONCERTO-3, prior relapsers to IFN-based therapy (relapsers); and CONCERTO-4, naïve, nonresponders, and relapsers. The eligibility criteria among the four studies were as follows: age between 20 and 70 years, chronic GT1 HCV infection, plasma HCV RNA levels ≥5.0 log10 IU/mL at screening, and no evidence of hepatic cirrhosis or hepatic failure. Dosage and treatment schedules in the CONCERTO-1,-2, and -3 studies were as follows: simeprevir 100 mg QD (53 CONCERTO-2 patients received this drug for 24 weeks) combined with PegIFN α-2a (180 μg once weekly) and RBV (total daily dose, 600–1000 mg/day, depending on body weight) for 12 weeks, followed by PegIFN α-2a and RBV alone for 12 weeks. PegIFN α-2b (1.5 μg/kg once weekly) was administered instead of PegIFN α-2a in CONCERTO-4. At week 24, patients either stopped or continued treatment with PegIFN and RBV according to response-guided therapy (RGT) criteria, except the CONCERTO-4 nonresponders. Patients who met the RGT criteria (i.e. achievement of HCV RNA levels <1.2 log10 IU/mL, either detectable or undetectable at week 4 and undetectable HCV RNA at week 12) could discontinue the therapy at week 24.
The primary endpoint of the original studies was SVR12, defined as the proportion of patients with undetectable HCV RNA at the end of treatment and at 12 weeks after the last dose of treatment, and the secondary endpoint was SVR24.
Because heterogeneity against SVR24 was strongly considered during prior treatment response, the dataset was divided into two subsets, naïve-relapsers and nonresponders, each of which was then individually analyzed. The analysis was first conducted using all integrated patient datasets, followed by the separate analysis of the two subsets divided based on prior treatment response.
Available parameters
SVR24 and viral relapse were used as outcome variables in this study. Although SVR12 was the primary endpoint for the CONCERTO studies, we used SVR24 as the outcome variable in this analysis because it is more commonly used as a primary efficacy parameter.
Body weight, height, hematology, blood chemistry, genotype of interleukin-28B (IL28B) and HCV GT1 amino acid substitutions in the NS3 region were assessed at baseline. Baseline blood samples were collected from patients who consented to undergo exploratory host DNA genotyping. The HCV NS3 region was genotyped by using standard population sequence. Inosine triphosphatase (ITPA) was genotyped by using the Invader Plus assay. Plasma HCV RNA quantification assay was performed from screening to week 72 (patients stopping PegIFN and RBV at week 24) by using the Roche COBAS® TaqMan® HCV Auto assay system (lower limit of quantification, 1.2 log10 IU/mL).
To select independent variables, we examined the following characteristics which were considered clinically relevant to SVR24 and viral relapse: previous treatment response (naïve patients, relapsers, nonresponders); age; sex; baseline body mass index (BMI, kg/m2); baseline HCV RNA level (log10 IU/mL); baseline platelet count (×109/L); baseline low density lipoprotein (LDL) cholesterol level (mmol/L); alpha-fetoprotein level (pmol/L); baseline fibrosis-4 (FIB-4) index (calculated as (age[year] × AST [U/L])/([PLT {10(9)/L}] × [ALT {U/L}](1/2)); baseline hemoglobin (g/dL); IL28B genotype (rs8099917; TT, TG, and GG); ITPA genotype (rs1127354; CC, CA, and AA); and plasma HCV RNA level (detectable or undetectable) at weeks 2 and 4.
Type of PegIFN, cumulative dose of simeprevir (mg/kg), minimum hemoglobin level (mg/dL), and RBV dose intensity [RBV-DI; calculated as cumulative dose of RBV (mg)/treatment period (week)/body surface area (m²)] were included to the analysis for nonresponders.
Furthermore, cumulative dose of RBV (g/kg) was included in the analysis of viral relapse in the nonresponders who treated ≥24 weeks.
Statistical analysis
Univariate and multivariate analyses were conducted to identify the potential predictors of SVR24 and viral relapse. Univariate analyses were sequentially performed for each independent variable to identify potential factors for further multivariate analysis. Fisher’s exact test and logistic regression models were used for comparisons between individual categorical independent variables and continuous variables, respectively. Variables with p values of <0.10 in the univariate analyses were included in the multivariate logistic regression analysis, which was performed using stepwise (backward) regression, and analyzed again for statistical significance. A p value of <0.05 was considered significant.
Receiver operating characteristic (ROC) curves were constructed for each logistic model, and the areas under the ROC (AUROC) curves were evaluated for predictive performance analysis. The 95 % confidence interval (CI) of the AUROC was computed by resampling 10,000 times (bootstrapping). The sensitivity and specificity of the ROC curves were determined at the cutoff point where (sensitivity + specificity − 1) it was maximal in the ROC curve (Youden index). ROC analysis was performed using the full model (including all statistically significant independent variables) as well as the limited model (each independent variable).
All statistical analyses were performed using R version 3.1.0 (a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/).
Results
Baseline demographics and clinical characteristics
In total, the CONCERTO studies enrolled 424 patients, of whom, 357 were eligible for this study. Specifically, 123, 106, 49, and 79 patients were from CONCERTO-1, -2, -3, and -4, respectively.
Baseline polymorphism on the HCV NS3 region was available from 353 patients, only five patients (1.4 %) presented Q80K and five patients (1.4 %) presented D168E polymorphism. The ITPA genotyping results were available for 308 patients. Patient demographics and baseline characteristics are summarized in Table 1.
Study medication
Table 2 presents information regarding completion and discontinuation of study treatment and results of efficacy endpoints for each subset. Treatment-naïve patients and relapsers were successfully treated with simeprevir and PegIFN plus RBV, with a treatment completion rate of 93.8 %. Except one patient, all patients completed treatment, met RGT criteria and finished study treatment at 24 weeks. On the other hand, the nonresponders exhibited less unfavorable completion rates, with 75.0 % of patients completing the planned treatment. Eighty-two (77.4 %) of 106 patients in CONCERTO-2 met RGT criteria, and 16 of 26 patients were treated ≥24 weeks of treatment in CONCERTO-4. The main reason for discontinuation (>5 %) was the virologic stopping criteria; 13.6 % of the patients discontinued the treatment because of these criteria.
Predictive factor analysis results
Integrated analysis of all patient datasets
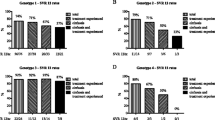
Eight variables revealed significant association with SVR24 in the integrated dataset of 357 patients using the univariate analyses. Subsequent multivariate analysis identified prior treatment responders and IL28B genotypes as significant predictors of baseline characteristics. The odds ratios (ORs) of prior treatment responses in naïve patients versus nonresponders and in relapsers versus nonresponders were 6.298 (95 % CI 3.149–13.192, p < 0.001) and 7.731 (95 % CI 3.011–22.845, p < 0.001), respectively (Table 3). The AUROC was 0.847 (95 % CI 0.798–0.889) in the full model (Fig. 1a).
ROC curves of full models in the (a) integrated dataset for SVR24 (n = 357), (b) subset of naïve patients and relapsers for SVR24 (n = 225), (c) subset of nonresponders for SVR24 (n = 132), and (d) subset of nonresponders who completed 24 weeks of treatment for viral relapse (n = 106). AUC areas under the curve, 95 % CI 95 % confidence interval
Subset analysis according to prior treatment response: naïve patients and relapsers
A total of 225 naïve patients and relapsers from CONCERTO-1, -3, and -4 were subjected to subset analysis. In the IL28B genotype, 70.7 % (159/225) of patients were TT genotype in this subset. The multivariate analysis showed that still the IL28B genotype was a baseline predictive factor. The OR of IL28B TT versus TG + GG was 4.106 (95 % CI 1.601–11.046, p = 0.004). Undetectable plasma HCV RNA at week 4 was identified as a treatment-associated variable, with an OR of 3.071 (95 % CI 1.089–8.241, p = 0.028; Table 4). Figure 1b shows the ROC curve of the full model, with an AUROC of 0.729 (95 % CI 0.6146–0.8327).
Subset analysis according to prior response: nonresponders
In total, 132 nonresponders from CONCERTO-2 and -4 were analyzed. IL28B genotype was more distinct in this subset than that in the naïve and relapser subset, i.e. 116 (87.9 %) of 132 patients exhibited TG and GG genotypes (Table 1). On univariate analysis, the IL28B genotype failed to show significance for prediction. Multivariate analysis revealed the ITPA genotype as a baseline predictive factor, with an OR of 2.506 (95 % CI 1.096–5.907, p = 0.003). SVR24 rate for ITPA CC was 36.0 % (27/75) and 57.9 % (22/38) for non-CC. Among 26 patients who discontinued the therapy within 24 weeks, 22 patients were available for ITPA genotype. Nineteen patients had ITPA CC and 18 of them relapsed. Prediction accuracy for ITPA was not favorable; AUROC was 0.598 (95 % CI 0.508–0.684). Undetectable plasma HCV RNA at week 4, as a treatment-associated variable, was identified as a significant predictive factor, with an OR of 3.333 (95 % CI 1.509–7.666, p = 0.004) (Table 5). Figure 1c shows the ROC curve of the full model, with an AUROC of 0.687 (95 % CI 0.5896–0.7784).
Predictive factors for viral relapse in prior nonresponders
In CONCERTO-2, although 82 of 106 patients had met the RGT criteria, 36 of them (43.9 %) relapsed after treatment completion. So, we analyzed predictive factors for viral relapse in nonresponders. A subset of 106 nonresponders from CONCERTO-2 and -4, who were treated with ≥24 weeks of therapy, were selected for this analysis. In this subset, a total of 51 patients (48.1 %) relapsed beyond 24 weeks of the therapy. On univariate analysis ITPA genotype failed to show significance for prediction. Relapse rates were 53.6 % (30/56) and 37.1 % (13/35) for ITPA CC and non-CC, respectively.
Multivariate analysis identified baseline HCV RNA level as a significant baseline factor, with an OR of 2.536 (95 % CI 1.100–6.322, p = 0.036); however, median RNA levels were mostly overlapped, 6.4 log10IU/mL (range 4.6–7.1) for patients with SVR and 6.5 log10IU/mL (range 5.5–7.3) for patients without SVR. Its prediction accuracy was not favorable, AUROC was 0.591 (95 % CI 0.4825–0.697). Plasma HCV RNA detection at week 4 and RBV-DI were identified as treatment-associated variables, with ORs of 2.922 (95 % CI 1.255–7.079, p = 0.015) and 0.327 (95 % CI 0.121–0.823, p = 0.021), respectively (Table 6). The AUROCs for RBV-DI and plasma HCV RNA level at week 4 were similar: 0.612 (95 % CI 0.502–0.718) and 0.618 (95 % CI 0.530–0.705), respectively. Figure 2 shows the box plot of RBV-DI. The RBV-DI threshold value was 2,790 mg/week/m² as per the ROC cutoff point calculations. The viral relapse rates were 57.1 % (95 % CI 44.2–70.1) and 38.0 % (95 % CI 24.5–51.5) in patients with RBV-DI below and above the threshold, respectively.
Figure 1d shows the ROC curve of the full model, with an AUROC of 0.707 (95 % CI 0.603–0.803).
Discussion
Similar to previous reports [11, 17], SVR24 rates were higher in naïve patients and relapsers than in nonresponders in the CONCERTO studies. Based on these findings, we primarily analyzed the integrated patient data of four CONCERTO studies and confirmed the heterogeneity of nonresponders with regard to other patient subsets.
Analysis of the naïve patients and relapsers revealed the IL28B genotype as a predictive factor for SVR24. IL28B has been a well-known predictor for PegIFN plus RBV in GT1 HCV patients [18, 19]. Its predictability has been reported in combination with telaprevir or boceprevir [20, 21]. According to these findings, even after significant improvements in SVR24 rates following the use of NS3/4A protease inhibitors, IL28B was clinically confirmed as an important predictive factor for SVR24 with IFN-containing regimens. Especially for naïve and relapsers with IL28B TT, shorter treatment duration by RGT criteria is desirable to reduce the potential treatment related AEs and treatment cost.
No factors related to hepatic status were identified in the present study. Platelet counts and alpha-fetoprotein levels have been previously identified [21, 22]. In the CONCERTO studies, patients were selected by the inclusion criteria. However, in real-world settings, a variety of patients are treated with simeprevir and PegIFN plus RBV. Therefore, further analysis using data from clinical settings is necessary to confirm the results of this study.
Considering the difficulties in achieving SVR in the nonresponders, determination of the predictive factors for SVR and viral relapse is critical. Basically, in contrast to naïve and relapsers, unfavorable IL28B genotype was dominant in this subset.
Mutation in the HCV RNA might be a potential reason for unsatisfactory response. Unfavorable SVR rate was reported in simeprevir-treated HCV genotype 1a patients with baseline Q80 K [23, 24]. Because patients in CONCERTO studies were mostly genotype 1b, prevalence of Q80K was only 1 % by population sequencing. Recently, Akuta et al. [25] studied a simeprevir-resistant variant with ultra-deep sequencing, such an approach could reveal new findings. Besides, the core region was not evaluated in the CONCERTO studies, which has been shown as a predictive factor for SVR with telaprevir and PegIFN plus RBV in nonresponders [22].
Several factors had to be considered with regard to treatment exposure. First, although pharmacokinetics of simeprevir was evaluated using population pharmacokinetics in the CONCERTO studies, no significant differences were observed across all studies (Table 2). Because 83.7–88.0 % of patients achieved complete early virologic response (cEVR) in CONCERTO-2, low SVR rates in the nonresponders might not have been caused by simeprevir adherence and its associated pharmacokinetics. Second, several studies have reported the development of an anti-IFN antibody in 5–15 % of patients, with higher rates of development in patients previously treated with IFN [26–28]. Anti-IFN antibody was not evaluated in the CONCERTO studies; it might cause treatment failure. Last, even after the advent of protease inhibitors, SVR24 rate was remarkably lower without RBV, and it has been considered as an essential component of protease inhibitor containing triple therapy [29]. The relationship between viral relapse and RBV dosage has been studied previously, adequate RBV-DI (≥6 mg/kg/day) was recommended in patients treated with PegIFN plus RBV for 48 weeks to prevent virus relapse [30, 31]. In the present study, relapse rate was statistically lower (38.0 %) in patients with RBV-DI > 2790 mg/week/m² (approximately 11 mg/kg/day). Both relapse rate and threshold of RBV-DI were higher in our study compared with those reported in previous studies, although patient background, particularly prior treatment response, and treatment duration were different among these studies. Bodeau et al. [32] studied the relationship between plasma concentrations of RBV at the end of treatment (week 48) and viral relapse, they reported that low RBV plasma concentrations resulted in significantly higher relapse rates. Overall, viral relapse could be mediated by RBV plasma concentrations even after the addition of a protease inhibitor to PegIFN plus RBV. Moreover, Oze et al. [33] have reported that the relapse rate was significantly decreased in patients with longer treatment durations (72 weeks) than in those with shorter treatment durations (48 weeks). Because most nonresponders completed the treatment at 24 weeks by the RGT criteria, a major limitation of this post hoc study was the lack of adequate evaluation of optimal treatment duration. Further investigations are warranted to explore and determine the optimal treatment duration with PegIFN plus RBV in nonresponders to prevent viral relapse after achievement of early viral response.
In addition, ITPA was revealed as a predictor of nonresponders, however, not in the ≥24 weeks treated nonresponders. Since ITPA CC was a well-known risk factor for RBV induced anemia, discontinuation of the therapy or insufficient RBV dose could be induced more frequently on this population. Patients with early discontinuation were mainly ITPA CC patients and they were removed from the viral relapse analysis; we considered this would make a discrepancy in the results of ITPA.
A few limitations to our study should be noted. Since the present study was a post hoc study which utilized merged data from registration studies and original studies were not designed to analyze predictive factors, it involved the selection of patients who were physically healthy, which would limit the extrapolation of our results in real-world clinical settings. Thus, additional studies using different participants, particularly in clinical settings, are warranted to confirm these results and their generalizability as well as to identify other meaningful predictive factors.
In conclusion, our results have identified the following clinically relevant predictive factors for SVR; undetectable plasma HCV RNA at week 4 for all patients and IL28B for naïve and relapsers. For viral relapse, RBV-DI and plasma HCV RNA detection were identified as clinically relevant predictive factors in nonresponders who were treated for ≥24 weeks without suboptimal response.
References
Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015;61:77–87
Hamada H, Yatsuhashi H, Yano K, et al. Impact of aging on the development of hepatocellular carcinoma in patients with post transfusion chronic hepatitis C. Cancer 2002;95:331–339
Schvarcz R, Weiland O, Wejstål R, et al. A randomized controlled open study of interferon alpha-2b treatment of chronic non-A, non-B posttransfusion hepatitis: no correlation of outcome to presence of hepatitis C virus antibodies. Scand J Infect Dis 1989;21:617–625
Omata M, Ito Y, Yokosuka O, et al. Randomized, double-blind, placebo-controlled trial of eight-week course of recombinant alpha-interferon for chronic non-A, non-B hepatitis. Dig Dis Sci 1991;36:1217–1222
Omata M, Kanda T, Yu M-L, et al. APASL consensus statements and management algorithms for hepatitis C virus infection. Hepatol Int 2012;6:409–435
European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C 2015. J Hepatol 2015;63(1):199-236; doi: 10.1016/j.jhep.2015.03.025
Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 2001;358:958–965
Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med 2002;347:975–982
Buti M, Mendez C, Schaper M, et al. Hepatitis C virus core antigen as a predictor of non-response in genotype 1 chronic hepatitis C patients treated with peginterferon alpha-2b plus ribavirin. J Hepatol 2004;40:527–532
Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 2010;138:1338–1345
Poynard T, Colombo M, Bruix J, et al. Peginterferon alfa-2b and ribavirin: effective in patients with hepatitis C who failed interferon alfa/ribavirin therapy. Gastroenterology 2009;136:1618–1628
Jacobson IM, McHutchison JG, Dusheiko G, et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med 2011;364:2405–2416
Zeuzem S, Andreone P, Pol S, et al. Telaprevir for retreatment of HCV infection. N Engl J Med 2011;364:2417–2428
Hayashi N, Izumi N, Kumada H, et al. Simeprevir with peginterferon/ribavirin for treatment-naïve hepatitis C genotype 1 patients in Japan: CONCERTO-1, a phase 3 trial. J Hepatol 2014;61:219–227
Izumi N, Hayashi N, Kumada H, et al. Once-daily simeprevir with peginterferon and ribavirin for treatment-experienced HCV genotype 1-infected patients in Japan: the CONCERTO-2 and CONCERTO-3 studies. J Gastroenterol 2014;49:941–953
Kumada H, Hayashi N, Izumi N, et al. Simeprevir (TMC435) once daily with peginterferon-α-2b and ribavirin in patients with genotype 1 hepatitis C virus infection: the CONCERTO-4 study. Hepatol Res 2015;45:501–513
Martinot-Peignoux M, Maylin S, Moucari R, et al. Virological response at 4 weeks to predict outcome of hepatitis C treatment with pegylated interferon and ribavirin. Antivir Ther 2009;14:501–511
Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009;461:399–401
McCarthy JJ, Li JH, Thompson A, et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology 2010;138:2307–2314
Akuta N, Suzuki F, Hirakawa M, et al. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology 2010;52:421–429
Hézode C, Fontaine H, Dorival C, et al. Effectiveness of telaprevir or boceprevir in treatment-experienced patients with HCV genotype 1 infection and cirrhosis. Gastroenterology 2014;147:132–142
Akuta N, Suzuki F, Seko Y, et al. Determinants of response to triple therapy of telaprevir, peginterferon, and ribavirin in previous non-responders infected with HCV genotype 1. J Med Virol 2012;84:1097–1105
Jacobson IM, Dore GJ, Foster GR, et al. Simeprevir with pegylated interferon alfa 2a plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-1): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet 2014;384:403–413
Manns M, Marcellin P, Poordad F, et al. Simeprevir with pegylated interferon alfa 2a or 2b plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-2): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 2014;384:414–426
Akuta N, Suzuki F, Sezaki H, et al. Evolution of simeprevir-resistant variants over time by ultra-deep sequencing in HCV genotype 1b. J Med Virol 2014;86:1314–1322
Leroy V, Baud M, de Traversay C, et al. Role of anti-interferon antibodies in breakthrough occurrence during alpha 2a and 2b therapy in patients with chronic hepatitis C. J Hepatol 1998;28:375–381
Matsuda F, Torii Y, Enomoto H, et al. Anti-interferon-α neutralizing antibody is associated with nonresponse to pegylated interferon-α plus ribavirin in chronic hepatitis C. J Viral Hepat 2012;19:694–703
Scagnolari C, Trombetti S, Soldà A, et al. Development and specificities of anti-interferon neutralizing antibodies in patients with chronic hepatitis C treated with pegylated interferon-α. Clin Microbiol Infect 2012;18:1033–1039
Hezode C, Forestier N, Dusheiko G, et al. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med 2009;360:1839–1850
Ogawa E, Furusyo N, Kajiwara E, et al. An inadequate dose of ribavirin is related to virological relapse by chronic hepatitis C patients treated with pegylated interferon alpha-2b and ribavirin. J Infect Chemother 2012;18:689–697
Kurosaki M, Hiramatsu N, Sakamoto M, et al. Age and total ribavirin dose are independent predictors of relapse after interferon therapy in chronic hepatitis C revealed by data mining analysis. Antivir Ther 2012;17:35–43
Bodeau S, Durand-Maugard C, Lemaire-Hurtel AS, et al. The end-of-treatment ribavirin concentration predicts hepatitis C virus relapse. Ther Drug Monit 2013;35:791–795
Oze T, Hiramatsu N, Yakushijin T, et al. The efficacy of extended treatment with pegylated interferon plus ribavirin in patients with HCV genotype 1 and slow virologic response in Japan. J Gastroenterol 2011;46:944–952
Acknowledgements
The authors would like to thank all patients and their families and the investigators and their staff at the following 45 study sites: Hokkaido University Hospital; Sapporo Kosei General Hospital; Tohoku University Hospital; Tokyo Medical University Ibaraki Medical Center; Saitama Medical University Hospital; Chiba University Hospital; Kohnodai Hospital; National Center for Global Health and Medicine; Showa University Hospital; Keio University Hospital; Kiyokawa Hospital; Juntendo University Hospital; Tokyo Medical And Dental University Hospital, Faculty of Medicine; Toranomon Hospital; Musashino Red Cross Hospital; Yokohama City University Medical Center; Showa University Fujigaoka Hospital; Toranomon Hospital, Kajigaya; Saiseikai Niigata Daini Hospital; Kanazawa University Hospital; University of Yamanashi Hospital; Shinshu University Hospital; Nagoya University Hospital; University Hospital, Kyoto Prefectural University of Medicine; Osaka City University Hospital; Osaka University Hospital; Osaka Koseinenkin Hospital; Ikeda Municipal Hospital; Osaka Rosai Hospital; Saiseikai Suita Hospital; Kinki University Hospital; Osaka City General Hospital; National Hospital Organization Osaka National Hospital; Kansai Rosai Hospital; The Hospital of Hyogo College of Medicine; Hiroshima University Hospital; Ehime University Hospital; Shin-Kokura Hospital; University Hospital of Occupational and Environmental Health; Kurume University Hospital; National Hospital Organization Nagasaki Medical Center; Kumamoto University Hospital; Kagoshima University Medical and Dental Hospital; The University of Tokyo Hospital; and Osaka Medical Center for Cancer and Cardiovascular Diseases. These studies were funded by Janssen Pharmaceutical K.K.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Masahiko Nakayama, Hisanori Kobayashi, Koji Fukushima, Miwako Ishido, Yuji Komada and Kazutake Yoshizawa are employees of Janssen Pharmaceutical K.K.
Informed consent in studies with human subjects
All patients provided written informed consent before study enrollment, and the study protocols conformed to Good Clinical Practice Guidelines and the ethical guidelines of the 1975 Declaration of Helsinki, as reflected in a priori approval by the human research committee of the respective institutions.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Nakayama, M., Kobayashi, H., Fukushima, K. et al. Predictive factors for 24 weeks sustained virologic response (SVR24) and viral relapse in patients treated with simeprevir plus peginterferon and ribavirin. Hepatol Int 10, 158–168 (2016). https://doi.org/10.1007/s12072-015-9654-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12072-015-9654-9