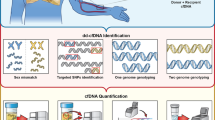
Abstract
Background
Circulating cell-free deoxyribonucleic acid (cfDNA) is promptly materializing as a highly useful tool for the surveillance of solid-organ transplant rejection. Donor-specific fraction (DF) cfDNA is a potential marker of selective donor organ injury. It is emerging as a promising analytical target in the near future. The aim of this systematic review is to throw light on the importance of cfDNA and future perspective in detecting acute rejection in heart transplantation.
Methods
An exhaustive search was carried out for this review article on the basis of literature available including scientific databases of PubMed, Embase, and ClinicalTrials.gov. The search engines were systematically explored using the search terms “cell free DNA,” “Heart transplant,” and “Rejection” from inception until August 2020, and narrative analysis was accomplished. Majority of the studies described endomyocardial biopsy-proven acute rejection as reference standard.
Results
After initial screening of 331 articles, 11 studies were included and discussed in detail in the present review article. Majority of the studies showed prospective designs. A firm correlation was noted between acute rejection (identified on endomyocardial biopsy) and cfDNA levels by most of the studies.
Conclusions
cfDNA is a promising tool to replace repeated biopsies to detect rejection. The development in the area of digital droplet polymerase chain reaction and massive parallel sequencing, along with the overall reduction in cost of sequencing with its automation, has helped establish its role in the transplant population.

Similar content being viewed by others
Data availability
All data used in the analysis are already available in the literature, and data sources are reported in the article.
References
Shah Z, Vuddanda V, Rali AS, Pamulapati H, Masoomi R, Gupta K. National trends and procedural complications from endomyocardial biopsy: results from the national inpatient sample, 2007–2014. Cardiology. 2018;141:125–31.
Saraiva F, Matos V, Goncalves L, Antunes M, Providencia LA. Complications of endomyocardial biopsy in heart transplant patients: a retrospective study of 2117 consecutive procedures. Transplant Proc. 2011;43:1908–12.
Fiorelli AI, Coelho GHB, Oliveira JL Jr, et al. Endomyocardial biopsy as risk factor in the development of tricuspid insufficiency after heart transplantation. Transplant Proc. 2009;41:935–7.
Patel JK, Kobashigawa JA. Should we be doing routine biopsy after heart transplantation in a new era of anti-rejection? Curr Opin Cardiol. 2006;21:127–31.
Stehlik J, Edwards LB, Kucheryavaya AY, et al. The Registry of the International Society for Heart and Lung Transplantation: twenty-seventh official adult heart transplant report--2010. J Heart Lung Transplant. 2010;29:1089–103.
Crespo-Leiro MG, Zuckermann A, Bara C, et al. Concordance among pathologists in the second Cardiac Allograft Rejection Gene Expression Observational Study (CARGO II). Transplantation. 2012;94:1172–7.
Yancy CW, Fonarow GC. Anticipating a new era in heart transplantation. JAMA Cardiol. 2020;5:625–7.
McCarthy JJ, McLeod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med. 2013;5:189 sr4.
Keating BJ, Pereira AC, Snyder M, Piening BD. Applying genomics in heart transplantation. Transpl Int. 2018;31:278–90.
North PE, Ziegler E, Mahnke DK, et al. Cell-free DNA donor fraction analysis in pediatric and adult heart transplant patients by multiplexed allele-specific quantitative PCR: validation of a rapid and highly sensitive clinical test for stratification of rejection probability. PLoS One. 2020;15:e0227385. https://doi.org/10.1371/journal.pone.0227385.
Beck J, Reinhard L, Oellerich M, et al. Early detection of rejection after heart transplantation by a universal digital PCR method. Clin Chem. 2015;61:S180.
De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77.
Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn. 2016;18:890–902.
Hidestrand M, Tomita-Mitchell A, Hidestrand PM, et al. Highly sensitive noninvasive cardiac transplant rejection monitoring using targeted quantification of donor-specific cell-free deoxyribonucleic acid. J Am Coll Cardiol. 2014;63:1224–6.
Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011;108:6229–34.
Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4:1.
Pös O, Biró O, Szemes T, Nagy B. Circulating cell-free nucleic acids: characteristics and applications. Eur J Hum Genet. 2018;26:937–45.
Kobashigawa JA, Khush K, Teuteberg J, et al. Initial analysis of the donor-derived cell-free DNA-outcomes allomap registry (D-OAR) study in heart transplant recipients undergoing surveillance for rejection. J Heart Lung Transplant. 2016;35:S33.
Richmond ME, Zangwill SD, Kindel SJ, et al. Donor fraction cell-free DNA and rejection in adult and pediatric heart transplantation. J Heart Lung Transplant. 2020;39:454–63.
Ragalie W, Stamm K, Hidestrand P, et al. Novel assay to calculate donor fraction of cell-free DNA in heart transplantation. J Am Coll Cardiol. 2018;71:A764.
Rodrigo ME, Agbor-Enoh S, Gorham S, et al. Association between LVAD and cardiac allograft injury following heart transplantation as assessed by cell-free DNA. J Heart Lung Transplant. 2017;36:S397.
Khush KK, Grskovic M, Luikart H, et al. Circulating cell-free DNA as a non-invasive marker of pediatric heart transplant rejection and immunosuppressive treatment. J Heart Lung Transplant. 2016;35:S75.
Crespo-Leiro MG, Stypmann J, Schulz U, et al. Clinical usefulness of gene-expression profile to rule out acute rejection after heart transplantation: CARGO II. Eur Heart J. 2016;37:2591–601.
Crespo-Leiro MG, Stypmann J, Schulz U, et al. Performance of gene-expression profiling test score variability to predict future clinical events in heart transplant recipients. BMC Cardiovasc Disord. 2015;15:120.
Agbor-Enoh S, Tunc I, De Vlaminck I, et al. Applying rigor and reproducibility standards to assay donor-derived cell-free DNA as a non-invasive method for detection of acute rejection and graft injury after heart transplantation. J Heart Lung Transplant. 2017;36:1004–12.
Valantine H, Shah P, Shah K, et al. Validation of donor-derived cell-free DNA to detect heart-transplant rejection. J Heart Lung Transplant. 2018;37:S78.
Khush KK, Patel J, Pinney S, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study. Am J Transplant. 2019;19:2889–99.
Home - ClinicalTrials.gov. [Internet]. [cited 2020 August 14] Available from: https://clinicaltrials.gov/ct2/show/NCT03477383?term=Donor+derived+cell+free+dna&cond=Heart+Transplant+Rejection&draw=2&rank=1. [Assessed 2020 August 8]
Home - ClinicalTrials.gov. [Internet]. [cited 2020 August 14] Available from: https://clinicaltrials.gov/ct2/show/NCT02178943?term=Donor+derived+cell+free+dna&cond=Heart+Transplant+Rejection&draw=2&rank=2 . [Assessed 2020 August 8]
Home - ClinicalTrials.gov. [Internet]. [cited 2020 August 14] Available from: https://clinicaltrials.gov/ct2/show/NCT03695601?term=Donor+derived+cell+free+dna&cond=heart+transplant+rejection&draw=2&rank=3. [Assessed 2020 August 8]
Dengu F. Next-generation sequencing methods to detect donor-derived cell-free DNA after transplantation. Transplant Rev. 2020;34:100542.
Pattar SK, Greenway SC. Circulating nucleic acids as biomarkers for allograft injury after solid organ transplantation: current state-of-the-art. Transpl Res Risk Manag. 2019;11:17–27.
What are single nucleotide polymorphisms (SNPs)? Available from: https://ghr.nlm.nih.gov/primer/genomicresearch/snp. [Assessed 2020 August 8]
Tucker T, Marra M, Friedman JM. Massively parallel sequencing: the next big thing in genetic medicine. Am J Hum Genet. 2009;85:142–54.
Li J, Dittmar RL, Xia S, et al. Cell-free DNA copy number variations in plasma from colorectal cancer patients. Mol Oncol. 2017;11:1099–111.
Wang Z, Zhang L, He L, et al. Low-depth whole genome sequencing reveals copy number variations associated with higher pathologic grading and more aggressive subtypes of lung non-mucinous adenocarcinoma. Chin J Cancer Res. 2020;32:334–46.
Hammer S, Meisner F, Dirschedl P, et al. Procalcitonin for differential diagnosis of graft rejection and infection in patients with heart and/or lung grafts. Intensive Care Med. 2000;26:S182–6.
Mena C, Wencker D, Krumholz HM, McNamara RL. Detection of heart transplant rejection in adults by echocardiographic diastolic indices: a systematic review of the literature. J Am Soc Echocardiogr. 2006;19:1295–300.
Vermes E, Pantaleon C, Auvet A, et al. Cardiovascular magnetic resonance in heart transplant patients: diagnostic value of quantitative tissue markers: T2 mapping and extracellular volume fraction, for acute rejection diagnosis. J Cardiovasc Magn Reson. 2018;20:59.
Patel PC, Hill DA, Ayers CR, et al. High-sensitivity cardiac troponin I assay to screen for acute rejection in patients with heart transplant. Circ Heart Fail. 2014;7:463–9.
Talha S, Charloux A, Enache I, Piquard F, Geny B. Mechanisms involved in increased plasma brain natriuretic peptide after heart transplantation. Cardiovasc Res. 2011;89:273–81.
Sparks JD, Boston U, Eghtesady P, Canter CE. B-type natriuretic peptide trends after pediatric heart transplantation. Pediatr Transplant. 2014;18:477–84.
Dieterlen MT, John K, Bittner HB, et al. Assessment of immunological biomarkers in the first year after heart transplantation. Dis Markers. 2015;2015:678061.
Oellerich M, Barten MJ, Armstrong VW. Biomarkers: the link between therapeutic drug monitoring and pharmacodynamics. Ther Drug Monit. 2006;28:35–8.
Crescioli C, Buonamano A, Scolletta S, et al. Predictive role of pretransplant serum CXCL10 for cardiac acute rejection. Transplantation. 2009;87:249–55.
Rotondi M, Netti GS, Lazzeri E, et al. High pretransplant serum levels of CXCL9 are associated with increased risk of acute rejection and graft failure in kidney graft recipients. Transpl Int. 2010;23:465–75.
Yamani MH, Taylor DO, Rodriguez ER, et al. Transplant vasculopathy is associated with increased AlloMap gene expression score. J Heart Lung Transplant. 2007;26:403–6.
Starling RC, Pham M, Valantine H, et al. Molecular testing in the management of cardiac transplant recipients: initial clinical experience. J Heart Lung Transplant. 2006;25:1389–95.
Gupta D, Bartra S, Shih R, et al. Correlation of Allomap scores in pediatric heart transplant recipients: are we ready to apply this to our patients? J Heart Lung Transplant. 2017;36:S267.
Pham MX, Teuteberg JJ, Kfoury AG, et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–900.
Melancon JK, Khalil A, Leman MJ. Donor derived cell free DNA: is it all the same. Kidney360. 2020;1:1118–23.
Van Huyen JPD, Tible M, Gay A, et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J. 2014;35:3194–202.
Tarazón E, Ortega A, Gil-Cayuela C, et al. SERCA2a: a potential non-invasive biomarker of cardiac allograft rejection. J Heart Lung Transplant. 2017;36:1322–8.
Camargo JF, Ahmed AA, Lindner MS, et al. Next-generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. F1000Res. 2019;8:1194.
Blauwkamp TA, Thair SA, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4:663–74.
Oellerich M, Schutz E, Kanzow P, et al. Use of graft-derived cell-free DNA as an organ integrity biomarker to reexamine effective tacrolimus trough concentrations after liver transplantation. Ther Drug Monit. 2014;36:136–40.
Lo YM, Tein MS, Pang CC, Yeung CK, Tong KL, Hjelm NM. Presence of donor-specific DNA in plasma of kidney and liver- transplant recipients. Lancet. 1998;351:1329–30.
Garcia Moreira V, Prieto Garcia B, Baltar Martin JM, Ortega Suárez F, Alvarez FV. Cell-free DNA as a noninvasive acute rejection marker in renal transplantation. Clin Chem. 2009;55:1958–66.
Wetterstrand KA. DNA sequencing costs: data from the NHGRI Genome Sequencing Program (GSP). Available at: www.genome.gov/sequencingcostsdata. Accessed [2020, August 11]. https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost.
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This manuscript represents a review article. Because this project involved no experimental design, Institutional Review Board approval was not required. Applicable EQUATOR Network (http://www.equator-network.org) guidelines were followed.
Registration of research studies
This prospective search was registered with the International Prospective Register of Systematic Reviews (PROSPERO; registration number CRD42020199218).
Statement of human and animal rights
Not required.
Informed consent
Not required.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sharma, D., Subramaniam, G., Sharma, N. et al. Cell-free DNA in the surveillance of heart transplant rejection. Indian J Thorac Cardiovasc Surg 37, 257–264 (2021). https://doi.org/10.1007/s12055-020-01130-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12055-020-01130-9