Abstract
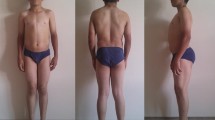
Myotonia congenita (MC) is a Mendelian inherited genetic disease caused by the mutations in the CLCN1 gene, encoding the main skeletal muscle ion chloride channel (ClC-1). The clinical diagnosis of MC should be suspected in patients presenting myotonia, warm-up phenomenon, a characteristic electromyographic pattern, and/or family history. Here, we describe the largest cohort of MC Spanish patients including their relatives (up to 102 individuals). Genetic testing was performed by CLCN1 sequencing and multiplex ligation-dependent probe amplification (MLPA). Analysis of selected exons of the SCN4A gene, causing paramyotonia congenita, was also performed. Mutation spectrum and analysis of a likely founder effect of c.180+3A>T was achieved by haplotype analysis and association tests. Twenty-eight different pathogenic variants were found in the CLCN1 gene, of which 21 were known mutations and seven not described. Gross deletions/duplications were not detected. Four probands had a pathogenic variant in SCN4A. Two main haplotypes were detected in c.180+3A>T carriers and no statistically significant differences were detected between case and control groups regarding the type of haplotype and its frequencies. A diagnostic yield of 51% was achieved; of which 88% had pathogenic variants in CLCN1 and 12% in SCN4A. The existence of a c.180+3A>T founder effect remains unsolved.


Similar content being viewed by others
References
Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P. et al. 2010 A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249.
Auton A., Abecasis G. R., Altshuler D. M., Durbin R. M., Bentley D. R., Chakravarti A. et al. 2015 A global reference for human genetic variation. Nature 526, 68–74.
Barrett J. C., Fry B., Maller J. and Daly M. J. 2005 Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265.
Becker P. E. 1979 Heterozygote manifestation in recessive generalized myotonia. Hum. Genet. 46, 325–329.
Berezin C., Glaser F., Rosenberg J., Paz I., Pupko T., Fariselli P. et al. 2004 ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 20, 1322–1324.
Brugnoni R., Kapetis D., Imbrici P., Pessia M., Canioni E., Colleoni L. et al. 2013 A large cohort of myotonia congenita probands: novel mutations and a high-frequency mutation region in exons 4 and 5 of the CLCN1 gene. J. Hum. Genet. 58, 581–587.
Chun S. and Fay J. C. 2009 Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561.
Colding-Jørgensen E. 2005 Phenotypic variability in myotonia congenita. Muscle Nerve 32, 19–34.
Consortium E. A., Lek M., Karczewski K., Minikel E., Samocha K., Banks E. et al. 2015 Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291.
Desmet F.-O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M. and Béroud C. 2009 Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67.
Dunø M. and Colding-Jørgensen E. 1993 Myotonia congenita, GeneReviews. University of Washington, Seattle (https://databases.lovd.nl/shared/variants/CLCN1/unique).
Dunø M. and Colding-Jørgensen E. 2005 Myotonia congénita, GeneReviews. University of Washington, Seattle (https://www.ncbi.nlm.nih.gov/books/NBK1355/).
Estévez R., Pusch M., Ferrer-Costa C., Orozco M. and Jentsch T. J. 2004 Functional and structural conservation of CBS domains from CLC chloride channels. J. Physiol. 557, 363–378.
Fialho D., Schorge S., Pucovska U., Davies N. P., Labrum R., Haworth A. et al. 2007 Chloride channel myotonia: exon 8 hot-spot for dominant-negative interactions. Brain 130, 3265–3274.
Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B. et al. 2002 The structure of haplotype blocks in the human genome. Science 296, 2225–2229.
Jentsch T. J., Poet M., Fuhrmann J. C. and Zdebik A. A. 2005 Physiological functions of Clc Cl-Channels gleaned from human genetic disease and mouse models. Annu. Rev. Physiol. 67, 779–807.
Jurkat-Rott K., Holzherr B., Fauler M. and Lehmann-Horn F. 2010 Sodium channelopathies of skeletal muscle result from gain or loss of function. Pflügers Arch. Eur. J. Physiol. 460, 239–248.
Koch M. C., Steinmeyer K., Lorenz C., Ricker K., Wolf F., Otto M. et al. 1992 The skeletal muscle chloride channel in dominant and recessive human myotonia. Science 257, 797–800.
Landrum M. J., Lee J. M., Benson M., Brown G., Chao C., Chitipiralla S. et al. 2015 ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862-D868.
Lehmann-Horn F. and Jurkat-Rott K. 1999 Voltage-gated ion channels and hereditary disease. Physiol. Rev. 79, 1317–1372.
Lehmann-Horn F., Jurkat-Rott K. and Rüdel R. 2008 Diagnostics and therapy of muscle channelopathies–guidelines of the Ulm muscle centre. Acta Myol. 27, 98–113.
Li B., Krishnan V. G., Mort M. E., Xin F., Kamati K. K., Cooper D. N. et al. 2009 Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25, 2744–2750.
Lipicky R. J., Bryant S. H. and Salmon J. H. 1971 Cable parameters, sodium, potassium, chloride, and water content, and potassium efflux in isolated external intercostal muscle of normal volunteers and patients with myotonia congenita. J. Clin. Invest. 50, 2091–2103.
Lossin C. and George A. L. 2008 Myotonia congenita. Adv. Genet. 63, 25–55.
Matthews E., Tan S. V, Fialho D., Sweeney M. G., Sud R. and Haworth A. 2008 What causes paramyotonia in the United Kingdom? Common and new SCN4A mutations revealed. Neurology 70, 50–53.
Mazón M. J., Barros F., De la Peña P., Quesada J. F., Escudero A., Cobo A. M. et al. 2012 Screening for mutations in Spanish families with myotonia. Functional analysis of novel mutations in CLCN1 gene. Neuromuscul. Disord. 22, 231–243.
Meyer-Kleine C., Steinmeyer K., Ricker K., Jentsch T. J. and Koch M. C. 1995 Spectrum of mutations in the major human skeletal muscle chloride channel gene (CLCN1) leading to myotonia. Am. J. Hum. Genet. 57, 1325–1334.
Papponen H., Toppinen T., Baumann P., Myllylä V., Leisti J., Kuivaniemi H. et al. 1999 Founder mutations and the high prevalence of myotonia congenita in northern Finland. Neurology 53, 297–302.
Pusch M. 2002 Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum. Mutat. 19, 423–434.
Reva B., Antipin Y. and Sander C. 2011 Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118.
Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J. et al. 2015 Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–423.
Schwarz J. M., Cooper D. N., Schuelke M. and Seelow D. 2014 MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362.
Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., Smigielski E. M. and Sirotkin K. 2001 dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311.
Simkin D. and Bendahhou S. 2011 Skeletal muscle na channel disorders. Front. Pharmacol. 2, 63. (https://doi.org/10.3389/fphar.2011.00063).
Skálová D., Zídková J., Voháňka S., Mazanec R., Mušová Z., Vondráček P. et al. 2013 CLCN1 Mutations in Czech patients with myotonia congenita, in silico analysis of novel and known mutations in the human dimeric skeletal muscle chloride channel. PLoS One 8, e82549.
Stenson P. D., Mort M., Ball E. V, Shaw K., Phillips A., Cooper D. N. et al. 2014 The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 133, 1–9.
Suetterlin K., Männikkö R. and Hanna M. G. 2014 Muscle channelopathies: recent advances in genetics, pathophysiology and therapy. Curr. Opin. Neurol. 27, 583–590.
Sun C., Tranebjaerg L., Torbergsen T., Holmgren G. and Van Ghelue M. 2001 Spectrum of CLCN1 mutations in patients with myotonia congenita in Northern Scandinavia. Eur. J. Hum. Genet. 9, 903–909.
Tennessen J. A., Bigham A. W., O’Connor T. D., Fu W., Kenny E. E., Gravel S. et al 2012 Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64–69.
Trip J., Drost G., Verbove D. J., van der Kooi A. J., Kuks J. B. M., Notermans N. C. et al. 2008 In tandem analysis of CLCN1 and SCN4A greatly enhances mutation detection in families with non-dystrophic myotonia. Eur. J. Hum. Genet. 16, 921–929.
Acknowledgements
We thank Ruben de Sancho and Amparo García Cardenal for their technical contribution in carrying out the experiments and Rocio Mena and Maria Victoria Gomez for capillary sequencing work. We also thank Will Brooks for improving the English language. This work was supported by a fellowship from the ‘Fundación J.L. Castaño’ in 2013 to C. Palma. CLCN1 gene homepage - Shared database [WWW Document], n.d. URL https://databases.lovd.nl/shared/genes/CLCN1(accessed4.18.18).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding Editor: H. A. Ranganath
Rights and permissions
About this article
Cite this article
Palma Milla, C., Prior De Castro, C., Gómez-González, C. et al. Myotonia congenita: mutation spectrum of CLCN1 in Spanish patients. J Genet 98, 71 (2019). https://doi.org/10.1007/s12041-019-1115-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-019-1115-0