Abstract
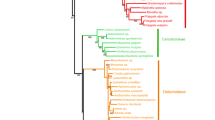
The families Lamiaceae and Verbenaceae comprise several closely related species that possess high morphological synapomorphic traits. Hence, there is a tendency of species misidentification using only the morphological characters. Herein, we evaluated the discriminatory power of the universal DNA barcodes (matK and rbcL) for 53 species spanning the two families. Using these markers, we inferred phylogenetic relationships and conducted species delimitation analysis using four delimitation methods: Automated Barcode Gap Discovery (ABGD), TaxonDNA, Bayesian Poisson Tree Processes (bPTP) and General Mixed Yule Coalescent (GMYC). The phylogenetic reconstruction based on the matK gene resolved the relationships between the families and further suggested the expansion of the Lamiaceae to include some core Verbanaceae genus, e.g., Gmelina. The rbcL marker using the TaxonDNA method displayed high species delimitation resolutions, while the ABGD, GMYC, and bPTP generated different number of Operational Taxonomic Units/genetic clusters. Our results underscored the efficiency of the matK and rbcL genes as reliable markers for resolving phylogenetic relationships and species delimitation of both families, respectively. The current study provides insights into the DNA barcode applications in these families, at the same time contributing to the current understanding of genetic divergence patterns in angiosperms.




Similar content being viewed by others
References
Angiosperm Phylogeny Group 2009 An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161 105–121
Abu-Asab MS and Cantino PD 1992 Pollen morphology in subfamily Lamioideae (Labiatae) and its phylogenetic implications; in Advances in Labiate Science (Eds.) RM Harley and H Reynolds (Royal Botanic Gardens, Kew, Richmond) pp. 97–112
Ashfaq M, Asif M, Anjum ZI and Zafar Y 2013 Evaluating the capacity of plant DNA barcodes to discriminate species of cotton (Gossypium: Malvaceae). Mol. Ecol. Resour. 13 574–582
Atkins S 2004 Verbenaceae; in The families and genera of flowering plants (Ed) JW Kadereit (Springer-Verlag, Berlin, Germany) pp. 449–468
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS and Donoghue MJ 1995 The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard. 82 247–277
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J and Sayers EW 2013 GenBank. Nucl. Acids Res. 41 D36–D42
Bos DH and Posada D 2015 Using models of nucleotide evolution to build phylogenetic trees. Dev. Comp. Immunol. 29 211–227
Bremer B, Bremer K, Heidari N, Erixon P, Olmstead RG, Anderberg AA, Kallersjo M and Barkhordarian E 2002 Phylogenetics of asterids based on 3 coding and 3 non-coding chloroplast DNA markers and the utility of non-coding DNA at higher taxonomic levels. Mol. Phyl. Evol. 24 274–301
Burgess K, Fazekas A J, Kesanakurti PR, Graham SW, Husband BC and Barrett SCH 2011 Discriminating plant species in a local temperate flora using the rbcL plus matK DNA barcode. Methods Ecol. Evol. 2 333–340
Cantino PD 1992 Evidence for a polyphyletic origin of the Labiatae. Ann. Mo. Bot. Gard. 79 361–379
Castresana J 2000 Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 540–552
CBOL 2009 Plant Working Group, A DNA barcode for land plants. Proc. Nat. Acad. Sci. USA 106 12794–12797
Chen J, Zhao J, Erickson DL, Xia N and Kress WJ 2015 Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol. Ecol. Resour. 15 337–348
Chen, Y P, Li B, Olmstead RG, Cantino PD, Liu ED and Xiang CL 2014 Phylogenetic placement of the enigmatic genus Holocheila (Lamiaceae) inferred from plastid DNA sequences. Taxon 63 355–366
Cowan RS and Fay MF 2012 Challenges in the DNA barcodingof plant material. Mol. Biol. 862 23–33
Cronquist A 1981 An integrated system of classification of flowering plants (Columbia University Press, New York)
Dahlgren R 1983 General aspects of angiosperm evolution and macrosystematics. Nordic J. Bot. 3 119–149
De Mattia F, Bruni I, Galimberti A, Cattaneo F, Casiraghi M and Labra M 2011 A comparative study of different DNA barcoding markers for the identification of some members of Lamiacaea. Food Res Int. 44 693–702
Dong W, Cheng T, Li C, Xu C, Long P and Zhou CCS 2014 Discriminating plants using the DNA barcode rbcLb:an appraisal based on a large data set. Mol. Ecol. Resour. 14 336–343
Doyle JJ, Doyle JL, Ballenger JA, Dickson EE, Kajita T and Ohashi H 1997 A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Am. J. Bot. 84 541–554
Dumas P, Barbut J, Le Ru B, Silvain JF, Clamens AL, et al 2015 Phylogenetic molecular species delimitations unravel potential new species in the pest genus Spodoptera GueneÂe, 1852 (Lepidoptera, Noctuidae). PLoS ONE 10 e0122407
Drummond AJ and Rambaut A 2007 BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7 214
Drummond AJ, Suchard MA, Xie D and Rambaut A 2012 Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29 1969–1973
Elansary HO, Ashfaq M, Ali HM and Yessoufou K 2017 The first initiative of DNA barcoding of ornamental plants from Egypt and potential applications in horticulture industry. PLoS ONE 12 e0172170
Ezard T, Fujisawa T and Barraclough TG 2009 splits: SPecies’ LImits by Threshold Statistics. R package version. 1.0–14/r31 (http://R-Forge.R-project.org/projects/splits/)
Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, Percy DM, Hajibabaei M and Barrett SC 2008 Multiple multilocus DNA barcodes from the plastid genome discriminate plantspecies equally well. PLoS ONE 3 e2802
Fazekas AJ, Kesanakurti PR, Burgess KS, Percy DM, Graham SW, Barrett SCH, Newmaster SG, Ibabaei MH and Husband BC 2009 Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Mol. Ecol. Resour. 9 130–139
Fu YM, Jiang W M and Fu CX 2011 Identification of species within Tetrastigma (Miq.) Planch. (Vitaceae) based on DNA barcoding techniques. J. Syst. Evol. 49 237–245
Fujisawa T and Barraclough TG 2013 Delimiting species using single-locus data and the generalized mixed yule coalescent approach: a revised method and evaluation on simulated data sets. Syst. Biol. 62 707–24
Gere J, Yessoufou K, Daru BH, Mankga LT, Maurin O and van der Bank M 2013 Incorporating trnH-psbA to the core DNA barcodes improves significantly species discrimination within southern African Combretaceae, in Z.T. Nagy, T. Backeljau, M. De Meyer, K. Jordaens (Eds) DNA barcoding: a practical tool for fundamental and applied biodiversity research. ZooKeys 365 127147
Gernhard T 2008 The conditioned reconstructed process. J. Theor. Biol. 253 769–778
Gonzalez MA, Baraloto C, Engel J, Mori SA, Pétronelli P, Riéra B, Roger A, Thébaud C and Chave J 2009 Identification of amazonian trees with DNA barcodes. PLoS ONE 4 e7483
Griffiths AJF, Miller H, Suzuki D T, Lewontin RC and Gelbart WM 2006 An Introduction to Genetic Analysis (New York: W. H. Freeman)
Guo YY, Huang LQ, Liu ZJ and Wang XQ 2016 Promise and Challenge of DNA Barcoding in Venus slipper (Paphiopedilum). PLoS ONE 11 e0146880
GRIN 2004 GRIN genera sometimes placed in Verbenaceae (Germplasm Resources Information Network: United States Department of Agriculture)
Hamilton CA, Hendrixson BE, Brewer MS and Bond JE 2014 An evaluation of sampling effects on multiple DNA barcoding methods leads to an integrative approach for delimiting species: a case study of the North American tarantula genus Aphonopelma (Araneae, Mygalomorphae, Theraphosidae). Mol. Phyl. Evol. 71 79–93
Hernández-León S, Little DP, Acevedo-Sandoval O, Gernandt DS, Rodríguez-Laguna R, Saucedo-García M, Arce-Cervantes O, Razo-Zárate R and Espitia-López J 2018 Plant core DNA barcode performance at a local scale: identification of the conifers of the state of Hidalgo, Mexico. Syst. Biod. 16 791–806
Hollingsworth PM, Graham SW and Little DP 2011 Choosing and using a Plant DNA barcode. PLoS ONE 6 e19254
Hutchinson J, Dalziel JM, Keay RWJ and Hepper FN 1963 Flora of West tropical Africa (Crown Agents for Oversea Governments and Administrations, London)
Jiang Y, Ding C, Zhang L, Yang R, Zhou Y and Tang L 2011 Identification of the genus Epimedium with DNA barcodes. J. Med. Plants Res. 5 6413–6417
Knowles LL and Carstens BC 2007 Delimiting species without monophyletic gene trees. Syst. Biol. 56 887–895
Kress WJ and Erickson DL 2007 A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2 e508
Kress WJ, Erickson DL, Andrew-Jones F, Swenson NG, Perez R, Sanjur O and Bermingham E, 2009 Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Nat. Acad. Sci. 106 18621–18626
Kress WJ, Erickson DL, Swenson NG, Thompson J, Uriarte M and Zimmerman JK 2010 Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican Forest Dynamics Plot. PLoS ONE 5 e15409
Kumar S and Gadagkar S R 2001 Disparity Index: a simple statistic to measure and test the homogeneity of substitution patterns between molecular sequences. Genetics 158 1321–1327
Kumar S, Stecher G and Tamura K 2016 MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874
Lahaye R, Savolainen V, Duthoit S, Maurin O and van der Bank M 2008 A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park (South Africa) as a model system. Nature Precedings. Available from http://hdl.handle.net/10101/npre.2008.1896.1. Accessed 2 Feb 2020
Lavin M, Herendeen PS and Wojciechowski MF 2005 Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst. Biol. 54 530–549
Li B, Cantino PD, Olmstead RG, Bramley GLC, Xiang CL, Ma ZH, Tan YH and Zhang DX 2016 A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci. Rep. 6 34343
Li HQ, Chen JY, Wang S and Xiong SZ 2012 Evaluation of six candidate DNA barcoding loci in Ficus (Moraceae) of China. Mol. Ecol. Resour. 12 783–790
Li S, Qian X, Zheng Z, Shi M, Change X, Li X, Liu J, Tu T and Zhang D 2018 DNA barcoding the flowering plants from the tropical coral islands of Xisha (China). Ecol. Evol. 8 10587–10593
Liu J, Moeller M, Gao LM, Zhang DQ and Li DZ 2011 DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol. Ecol. Res. 11 89–100
Ma PF, Zhang YX, Zeng CX, Guo ZH and Li DZ 2014 Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst. Biol. 63 933–950
Marx HE, O’Leary N, Yuan Y, Lu-Irving P, Tank DC, Mulgara ME and Olmstead RG 2010 A molecular phylogeny and classification of Verbenaceae. Am. J. Bot. 97 1647–1663
Meier R, Kwong S, Vaidya G and Ng P K L 2006 DNA Barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst. Biol. 55 715–728
Miller MA, Pfeiffer W and Schwartz T 2010 Creating the CIPRES science gateway for inference of large phylogenetic trees, in Proceedings of the Gateway Computing Environments Workshop, New Orleans, LA. pp. 1–8
Nee S 2006 Birth-death models in macroevolution. Annu. Rev. Ecol. Evol. Syst. 37 1–17
Nicolalde-Morejón F, Vergara-Silva F, González-Astorga J and Stevenson D W 2010 Character-based, population-level DNA barcoding in Mexican species of Zamia L. (Zamiaceae: Cycadales). Mitochondrial DNA 21 51–59
Olmstead RG, dePamphilis CW, Wolfe AD, Young ND, Elisens WJ and Reeves PA 2001 Disintegration of the Scrophulariaceae. Am. J. Bot. 88 348–361
Olmstead RG, Jansen RK, Kim KJ and Wagstaff SJ 2000 The phylogeny of the Asteridae s.l. based on chloroplast ndhF sequences. Mol. Phyl. Evol. 16 96–112
Oxelman B, Kornhall P, Olmstead RG and Bremer B 2005 Further disintegration of the Scrophulariaceae. Taxon 54 411–425
Paradis E, Claude J and Strimmer K 2004 APE: analyses of phylogenetics and evolution in R language. Bioinfomatics 20 289–290
Parmentier I, Duminil J, Kuzmina M, Philippe M, Thomas D W, Kenfack D, Chuyong GB, Cruaud C and Hardy OJ 2013 How effective are DNA barcodes in the identification of African rainforest trees? PLoS ONE 8 e54921
Pennisi E 2007 Taxonomy. Wanted: A barcode for plants. Science 318 190–191
Pollock D, Zwick D J, McGuire JA and Hillis D M 2002 Increased taxon sampling is advantageous for phylogenetic inference. Syst. Biol. 51 664–671
Posada D and Buckley TR 2004 Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53 793–808
Puillandre N, Lambert A, Brouillet S and Achaz G 2011 ABGD, Automatic barcode discovery for primary species delimitation. Mol. Ecol. 21 1864–1877
R Core Team 2012 R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Rambaut A and Drummond A J 2009 Tracer version 1.5 [computer program] (http://beast.bio.ed.ac.uk)
Rambaut A and Drummond A J 2010. TreeAnnotator version 1.6.1 [computer program] (http://beast.bio.ed.ac.uk)
Rambaut A 2014 FigTree v1.4.2, a graphical viewer of phylogenetic trees (http://tree.bio.ed.ac.uk/software/figtree/)
Refulio-Rodriguez N F and Olmstead R G 2014 Phylogeny of Lamiidae. Am. J. Bot. 101 287–299
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA and Huelsenbeck JP 2012 MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 539–542
Rozas J 2009 Polymorphism Analysis using DnaSP, in Bioinformatics for DNA Sequence Analysis: Methods in Molecular Biology Series (Ed) Posada D (Humana Press NJ USA) pp. 337–350
Ryding O 1995 Pericarp structure and phylogeny of the Lamiaceae-Verbenaceae-complex. Plant Syst. Evol. 198 101–141
Saarela JM, Sokoloff PC, Gillespie LJ, Consaul LL and Bull RD 2013 DNA barcoding the Canadian Arctic flora: core plastid barcodes (rbcL + matK) for 490 vascular plant species. PLoS ONE 8 e77982
Saddhe AA, Jamdade RA, and Kumar K 2017 Evaluation of multilocus marker efficacy for delineating mangrove species of West Coast India. PLoS ONE 12 e0183245
Sahu S K, Singh R, and Kathiresan K, 2016 Multi-gene phylogenetic analysis reveals the multiple origin and evolution of mangrove physiological traits through exaptation, Estuarine, Coastal and Shelf. Science 183 41–51
Sanders R W 2001 The genera of Verbenaceae in the southeastern United States. Harvard Papers Bot. 5 303–358
Schäferhoff B, Fleischmann A, Fischer E, Albach DC, Borsch T, Heubl G and Müller KF 2010 Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences. BMC Evol. Biol. 10 352
Scheen A, Bendiksby M, Ryding O, Mathiesen C, Albert VA and Lindqvist C 2010 Molecular phylogenetics, character evolution, and suprageneric classification of Lamioideae (Lamiaceae). Ann. Mo. Bot. Gard. 97 191–217
Selvaraj D, Sarma RK, and Sathishkumar R 2008 Phylogenetic analysis of chloroplast matK gene from Zingiberaceae for plant DNA barcoding. Bioinformation 3 24–27
Stamatakis A 2006 RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 2688–2690
Stoeckle MY, Gamble CC, Kirpekar R, Young G, Ahmed S and Little D P 2011 Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci. Rep. 1 42
Surya SD, Swati S, and Partadeb G 2014 Phylogenetic relationships among three species of the mangrove genus Avicennia found in Indian Sundarban, as revealed by RAPD analysis. Asian J. Plant Sci. 4 25–30
Swofford DL 2002 PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4b10. Sinauer Associates, Sunderland, Massachusetts, USA
Talavera G and Castresana J 2007 Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56 564–577
Tang CG, Humphreys AM, Fontaneto D and Barraclough TG 2014 Effects of phylogenetic reconstruction method on the robustness of species delimitation using single-locus data. Methods Ecol. Evol. 5 1086–1094
Thorne RF 1983 Proposed new realignments in the angiosperms. Nordic J. Bot. 3 85–117
Vinitha RM, Kumar SU, Aishwarya K, Sabu M and Thomas G 2014 Prospects for discriminating Zingiberaceae species in India using DNA barcodes. J. Integr Plant Biol. 56 760–773
Wagstaff SJ and Olmstead RG 1997 Phylogeny of the Labiatae and Verbenaceae inferred from rbcL sequences. Syst. Bot. 22 165–179
Wagstaff SJ, Hickerson L, Spangler R, Reeves PA and Olmstead R G 1998 Phylogeny in Labiatae s.l., inferred from cpDNA sequences. Plant Syst. Evol. 209 265–274
Weiss M, Macher J N, Seefeldt MA, and Leese F 2014 Molecular evidence for further overlooked species within the Gammarus fossarum complex (Crustacea: Amphipoda). Hydrobiologia 721 165–184
Xiang XG, Hu H, Wang W and Jin XH 2011 DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): a test of DNA barcode candidates. Mol. Ecol. Resour. 11 1012–1021
Yan LJ, Liu J, Möller M, Zhang L, Zhang XM, Li DZ and Gao LM 2015 DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the Himalaya-Hengduan Mountains. Mol. Ecol. Resour. 15 932–944
Yuan YW, Mabberley DJ, Steane DA and Olmstead RG 2010 Further disintegration and redefinition of Clerodendrum (Lamiaceae): implications for the understanding of the evolution of an intriguing breeding strategy. Taxon 59 125–133
Zhang CY, Wang FY, Yan HF, Hao G and Hu CM 2012 Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol. Ecol. Resour. 12 98–108
Zhang J, Kapli P and Pavlidis P 2013 A. Stamatakis, A General species delimitation method with applications to phylogenetic placements. Bioinformatics 29 2869–2876
Zhang J, Chen, M, Dong X, Lin, R, Fan J and Chen Z 2015 Evaluation of four commonly used DNA barcoding loci for chinese medicinal plants of the family Schisandraceae. PLoS ONE 10 e0125574
Zou S, Fei C, Song J, Bao Y, He M, and Wang C 2016 Combining and comparing coalescent, distance and character-based approaches for barcoding microalgaes: a test with Chlorella-like species (Chlorophyta). PLoS ONE 11 e0153833
Acknowledgements
The authors gratefully acknowledge Dr. Nneji Lotanna Micah of the Kunming Institute of Zoology, Chinese Academy of Sciences, China, and all those who have contributed towards the success of this research. Also, we appreciate the intellectual guidance of the anonymous reviewers that improved the quality of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by NG Prasad.
Corresponding editor: NG Prasad
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Oyebanji, O.O., Chukwuma, E.C., Bolarinwa, K.A. et al. Re-evaluation of the phylogenetic relationships and species delimitation of two closely related families (Lamiaceae and Verbenaceae) using two DNA barcode markers. J Biosci 45, 96 (2020). https://doi.org/10.1007/s12038-020-00061-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-020-00061-2