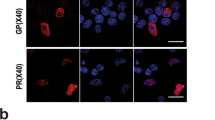
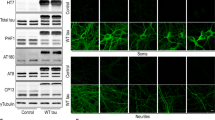
Abstract
The cellular prion protein (PrPC) is a ubiquitous glycoprotein highly expressed in the brain where it is involved in neurite outgrowth, copper homeostasis, NMDA receptor regulation, cell adhesion, and cell signaling. Conformational conversion of PrPC into its insoluble and aggregation-prone scrapie form (PrPSc) is the trigger for several rare devastating neurodegenerative disorders, collectively referred to as prion diseases. Recent work indicates that the ubiquitin–proteasome system is involved in quality control of PrPC. To better dissect the role of ubiquitination in PrPC physiology, we focused on the E3 RING ubiquitin ligase tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6). Here, we report that PrPC interacts with TRAF6 both in vitro, in cells, and in vivo, in the mouse brain. Transient overexpression of TRAF6 indirectly modulates PrPC ubiquitination and triggers redistribution of PrPC into the insoluble fraction. Importantly, in the presence of wild-type TRAF6, but not a mutant lacking E3 ligase activity, PrPC accumulates into cytoplasmic aggresome-like inclusions containing TRAF6 and p62/SQSTM1. Our results suggest that TRAF6 ligase activity could exert a role in the regulation of PrPC redistribution in cells under physiological conditions. This novel interaction may uncover possible mechanisms of cell clearance/reorganization in prion diseases.





Similar content being viewed by others
Availability of Data and Materials
Data and materials used in this study and supporting the conclusions of this article are included within the article.
Code Availability
Not applicable.
Abbreviations
- PrPC :
-
Cellular prion protein
- PrPSc :
-
Scrapie prion protein
- TRAF6:
-
Tumor necrosis factor receptor-associated factor 6
- DN:
-
Deleted of the N terminus
References
Stahl N, Borchelt DR, Hsiao K, Prusiner SB (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51:229–240
Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG (1983) Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 35:349–358
Bockman JM, Kingsbury DT, McKinley MP, Bendheim PE, Prusiner SB (1985) Creutzfeldt-Jakob disease prion proteins in human brains. N Engl J Med 312:73–78. https://doi.org/10.1056/NEJM198501103120202
Zanusso G, Petersen RB, Jin T, Jing Y, Kanoush R, Ferrari S, Gambetti P, Singh N (1999) Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J Biol Chem 274:23396–23404
Mishra RS, Bose S, Gu Y, Li R, Singh N (2003) Aggresome formation by mutant prion proteins: the unfolding role of proteasomes in familial prion disorders. J Alzheimers Dis 5:15–23
Laszlo L, Lowe J, Self T, Kenward N, Landon M, McBride T, Farquhar C, McConnell I et al (1992) Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J Pathol 166:333–341. https://doi.org/10.1002/path.1711660404
Ironside JW, McCardle L, Hayward PA, Bell JE (1993) Ubiquitin immunocytochemistry in human spongiform encephalopathies. Neuropathol Appl Neurobiol 19:134–140
Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, Clarke AR, van Leeuwen FW et al (2007) Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell 26:175–188. https://doi.org/10.1016/j.molcel.2007.04.001
McKinnon C, Goold R, Andre R, Devoy A, Ortega Z, Moonga J, Linehan JM, Brandner S et al (2016) Prion-mediated neurodegeneration is associated with early impairment of the ubiquitin-proteasome system. Acta Neuropathol 131:411–425. https://doi.org/10.1007/s00401-015-1508-y
Kang SC, Brown DR, Whiteman M, Li R, Pan T, Perry G, Wisniewski T, Sy MS et al (2004) Prion protein is ubiquitinated after developing protease resistance in the brains of scrapie-infected mice. J Pathol 203:603–608. https://doi.org/10.1002/path.1555
Yedidia Y, Horonchik L, Tzaban S, Yanai A, Taraboulos A (2001) Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J 20:5383–5391. https://doi.org/10.1093/emboj/20.19.5383
Shao J, Choe V, Cheng H, Tsai YC, Weissman AM, Luo S, Rao H (2014) Ubiquitin ligase gp78 targets unglycosylated prion protein PrP for ubiquitylation and degradation. PLoS ONE 9:e92290. https://doi.org/10.1371/journal.pone.0092290
Lee JH, Han Y-S, Yoon YM, Yun CW, Yun SP, Kim SM, Kwon HY, Jeong D et al (2017) Role of HSPA1L as a cellular prion protein stabilizer in tumor progression via HIF-1α/GP78 axis. Oncogene 36(47):6555–6567. https://doi.org/10.1038/onc.2017.263
Homma T, Ishibashi D, Nakagaki T, Fuse T, Mori T, Satoh K, Atarashi R, Nishida N (2015) Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Sci Rep 5:11028. https://doi.org/10.1038/srep11028
Gimenez AP, Richter LM, Atherino MC, Beirao BC, Favaro C Jr, Costa MD, Zanata SM, Malnic B et al (2015) Identification of novel putative-binding proteins for cellular prion protein and a specific interaction with the STIP1 homology and U-Box-containing protein 1. Prion 9:355–366. https://doi.org/10.1080/19336896.2015.1075347
Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV (1996) TRAF6 is a signal transducer for interleukin-1. Nature 383:443–446. https://doi.org/10.1038/383443a0
Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ (2011) MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146:448–461. https://doi.org/10.1016/j.cell.2011.06.041
Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ (2013) MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife 2:e00785. https://doi.org/10.7554/eLife.00785
Geetha T, Jiang J, Wooten MW (2005) Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol Cell 20:301–312. https://doi.org/10.1016/j.molcel.2005.09.014
Khursigara G, Orlinick JR, Chao MV (1999) Association of the p75 neurotrophin receptor with TRAF6. J Biol Chem 274:2597–2600
Ma Q, Ruan H, Peng L, Zhang M, Gack MU, Yao WD (2017) Proteasome-independent polyubiquitin linkage regulates synapse scaffolding, efficacy, and plasticity. Proc Natl Acad Sci U S A 114:E8760–E8769. https://doi.org/10.1073/pnas.1620153114
Vilotti S, Codrich M, Dal Ferro M, Pinto M, Ferrer I, Collavin L, Gustincich S, Zucchelli S (2012) Parkinson’s disease DJ-1 L166P alters rRNA biogenesis by exclusion of TTRAP from the nucleolus and sequestration into cytoplasmic aggregates via TRAF6. PLoS ONE 7:e35051. https://doi.org/10.1371/journal.pone.0035051
Zucchelli S, Marcuzzi F, Codrich M, Agostoni E, Vilotti S, Biagioli M, Pinto M, Carnemolla A et al (2011) Tumor necrosis factor receptor-associated factor 6 (TRAF6) associates with huntingtin protein and promotes its atypical ubiquitination to enhance aggregate formation. J Biol Chem 286:25108–25117. https://doi.org/10.1074/jbc.M110.187591
Zucchelli S, Codrich M, Marcuzzi F, Pinto M, Vilotti S, Biagioli M, Ferrer I, Gustincich S (2010) TRAF6 promotes atypical ubiquitination of mutant DJ-1 and alpha-synuclein and is localized to Lewy bodies in sporadic Parkinson’s disease brains. Hum Mol Genet 19:3759–3770. https://doi.org/10.1093/hmg/ddq290
Giachin G, Mai PT, Tran TH, Salzano G, Benetti F, Migliorati V, Arcovito A, Della Longa S et al (2015) The non-octarepeat copper binding site of the prion protein is a key regulator of prion conversion. Sci Rep 5:15253. https://doi.org/10.1038/srep15253
van den Ent F, Lowe J (2006) RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67:67–74. https://doi.org/10.1016/j.jbbm.2005.12.008
Didonna A, Venturini AC, Hartman K, Vranac T, CurinSerbec V, Legname G (2015) Characterization of four new monoclonal antibodies against the distal N-terminal region of PrP(c). PeerJ 3:e811. https://doi.org/10.7717/peerj.811
Petsch B, Muller-Schiffmann A, Lehle A, Zirdum E, Prikulis I, Kuhn F, Raeber AJ, Ironside JW et al (2011) Biological effects and use of PrPSc- and PrP-specific antibodies generated by immunization with purified full-length native mouse prions. J Virol 85:4538–4546. https://doi.org/10.1128/JVI.02467-10
Barry RA, Prusiner SB (1986) Monoclonal antibodies to the cellular and scrapie prion proteins. J Infect Dis 154:518–521
Rane NS, Chakrabarti O, Feigenbaum L, Hegde RS (2010) Signal sequence insufficiency contributes to neurodegeneration caused by transmembrane prion protein. J Cell Biol 188:515–526. https://doi.org/10.1083/jcb.200911115
Rane NS, Kang SW, Chakrabarti O, Feigenbaum L, Hegde RS (2008) Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev Cell 15:359–370. https://doi.org/10.1016/j.devcel.2008.06.015
Chakrabarti O, Ashok A, Hegde RS (2009) Prion protein biosynthesis and its emerging role in neurodegeneration. Trends Biochem Sci 34:287–295. https://doi.org/10.1016/j.tibs.2009.03.001
Rane NS, Yonkovich JL, Hegde RS (2004) Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J 23:4550–4559. https://doi.org/10.1038/sj.emboj.7600462
Ma J, Wollmann R, Lindquist S (2002) Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science 298:1781–1785. https://doi.org/10.1126/science.1073725
Grenier C, Bissonnette C, Volkov L, Roucou X (2006) Molecular morphology and toxicity of cytoplasmic prion protein aggregates in neuronal and non-neuronal cells. J Neurochem 97:1456–1466. https://doi.org/10.1111/j.1471-4159.2006.03837.x
Ma J, Lindquist S (2002) Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science 298:1785–1788. https://doi.org/10.1126/science.1073619
Zapata JM, Pawlowski K, Haas E, Ware CF, Godzik A, Reed JC (2001) A diverse family of proteins containing tumor necrosis factor receptor-associated factor domains. J Biol Chem 276:24242–24252. https://doi.org/10.1074/jbc.M100354200
Force WR, Glass AA, Benedict CA, Cheung TC, Lama J, Ware CF (2000) Discrete signaling regions in the lymphotoxin-beta receptor for tumor necrosis factor receptor-associated factor binding, subcellular localization, and activation of cell death and NF-kappaB pathways. J Biol Chem 275:11121–11129
Hostager BS, Catlett IM, Bishop GA (2000) Recruitment of CD40 and tumor necrosis factor receptor-associated factors 2 and 3 to membrane microdomains during CD40 signaling. J Biol Chem 275:15392–15398. https://doi.org/10.1074/jbc.M909520199
Ha H, Kwak HB, Lee SK, Na DS, Rudd CE, Lee ZH, Kim HH (2003) Membrane rafts play a crucial role in receptor activator of nuclear factor kappaB signaling and osteoclast function. J Biol Chem 278:18573–18580. https://doi.org/10.1074/jbc.M212626200
Kim SJ, Hegde RS (2002) Cotranslational partitioning of nascent prion protein into multiple populations at the translocation channel. Mol Biol Cell 13:3775–3786. https://doi.org/10.1091/mbc.E02-05-0293
Fons RD, Bogert BA, Hegde RS (2003) Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol 160:529–539. https://doi.org/10.1083/jcb.200210095
Satpute-Krishnan P, Ajinkya M, Bhat S, Itakura E, Hegde RS, Lippincott-Schwartz J (2014) ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell 158:522–533. https://doi.org/10.1016/j.cell.2014.06.026
Chakrabarti O, Rane NS, Hegde RS (2011) Cytosolic aggregates perturb the degradation of nontranslocated secretory and membrane proteins. Mol Biol Cell 22:1625–1637. https://doi.org/10.1091/mbc.E10-07-0638
Schultheiss U, Puschner S, Kremmer E, Mak TW, Engelmann H, Hammerschmidt W, Kieser A (2001) TRAF6 is a critical mediator of signal transduction by the viral oncogene latent membrane protein 1. EMBO J 20:5678–5691. https://doi.org/10.1093/emboj/20.20.5678
Lamothe B, Campos AD, Webster WK, Gopinathan A, Hur L, Darnay BG (2008) The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL. J Biol Chem 283:24871–24880. https://doi.org/10.1074/jbc.M802749200
Lee JT, Wheeler TC, Li L, Chin LS (2008) Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum Mol Genet 17:906–917. https://doi.org/10.1093/hmg/ddm363
Polekhina G, House CM, Traficante N, Mackay JP, Relaix F, Sassoon DA, Parker MW, Bowtell DD (2002) Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat Struct Biol 9:68–75. https://doi.org/10.1038/nsb743
Johnston JA, Ward CL, Kopito RR (1998) Aggresomes: a cellular response to misfolded proteins. J Cell Biol 143:1883–1898
Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, Fischbeck KH (2003) Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet 12:749–757. https://doi.org/10.1093/hmg/ddg074
Zucchelli S, Vilotti S, Calligaris R, Lavina ZS, Biagioli M, Foti R, De Maso L, Pinto M et al (2009) Aggresome-forming TTRAP mediates pro-apoptotic properties of Parkinson’s disease-associated DJ-1 missense mutations. Cell Death Differ 16:428–438. https://doi.org/10.1038/cdd.2008.169
Olzmann JA, Chin LS (2008) Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy 4:85–87
Kovacs GG, Molnar K, Keller E, Botond G, Budka H, Laszlo L (2012) Intraneuronal immunoreactivity for the prion protein distinguishes a subset of E200K genetic from sporadic Creutzfeldt-Jakob Disease. J Neuropathol Exp Neurol 71:223–232. https://doi.org/10.1097/NEN.0b013e318248aa70
Babu JR, Geetha T, Wooten MW (2005) Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem 94:192–203. https://doi.org/10.1111/j.1471-4159.2005.03181.x
Kang SW, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS (2006) Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127:999–1013. https://doi.org/10.1016/j.cell.2006.10.032
Homma T, Ishibashi D, Nakagaki T, Satoh K, Sano K, Atarashi R, Nishida N (2014) Increased expression of p62/SQSTM1 in prion diseases and its association with pathogenic prion protein. Sci Rep 4:4504. https://doi.org/10.1038/srep04504
Kim SJ, Rahbar R, Hegde RS (2001) Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J Biol Chem 276:26132–26140. https://doi.org/10.1074/jbc.M101638200
Inoue J, Ishida T, Tsukamoto N, Kobayashi N, Naito A, Azuma S, Yamamoto T (2000) Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Exp Cell Res 254:14–24. https://doi.org/10.1006/excr.1999.4733
Wajant H, Henkler F, Scheurich P (2001) The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal 13:389–400
Ha H, Han D, Choi Y (2009) TRAF-mediated TNFR-family signaling. Curr Protoc Immunol Chapter 11(Unit11):9D. https://doi.org/10.1002/0471142735.im1109ds87
Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG (2007) Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem 282:4102–4112. https://doi.org/10.1074/jbc.M609503200
Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ (2009) Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461:114–119. https://doi.org/10.1038/nature08247
Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A et al (1996) Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci U S A 93:14945–14949
Xing Y, Yao X, Li H, Xue G, Guo Q, Yang G, An L, Zhang Y et al (2017) Cutting Edge: TRAF6 mediates TLR/IL-1R signaling-induced nontranscriptional priming of the NLRP3 inflammasome. J Immunol 199:1561–1566. https://doi.org/10.4049/jimmunol.1700175
Ardley HC, Scott GB, Rose SA, Tan NG, Robinson PA (2004) UCH-L1 aggresome formation in response to proteasome impairment indicates a role in inclusion formation in Parkinson’s disease. J Neurochem 90:379–391. https://doi.org/10.1111/j.1471-4159.2004.02485.x
Ma S, Attarwala IY, Xie XQ (2019) SQSTM1/p62: a potential target for neurodegenerative disease. ACS Chem Neurosci 10:2094–2114. https://doi.org/10.1021/acschemneuro.8b00516
Acknowledgements
This work is dedicated to the memory of our beloved friend and colleague Silvia Zucchelli who passed away in 2019 during the writing and reviewing of this manuscript.
The authors wish also to thank Drs Ilaria Poggiolini and Thao Mai Phuong for their involvement in the early stages of the project.
Funding
This work was supported by SISSA intramural funding to GL.
Author information
Authors and Affiliations
Contributions
LM and MC designed and performed the experiments, analyzed the data, and wrote the manuscript; FP and SG analyzed the data and the manuscript; SZ designed the experiments, analyzed the results, and wrote the manuscript; GL designed the experiments, analyzed the results, and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
All animal experiments were performed in accordance with European guidelines for animal care (European Community Council Directive, November 24, 1986 86/609/EEC) and following Italian Board of Health permissions (Law n. 116/1992).
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Silvia Zucchelli was Deceased (April 28, 1972 - October 14, 2019)
Rights and permissions
About this article
Cite this article
Masperone, L., Codrich, M., Persichetti, F. et al. The E3 Ubiquitin Ligase TRAF6 Interacts with the Cellular Prion Protein and Modulates Its Solubility and Recruitment to Cytoplasmic p62/SQSTM1-Positive Aggresome-Like Structures. Mol Neurobiol 59, 1577–1588 (2022). https://doi.org/10.1007/s12035-021-02666-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02666-6