Abstract
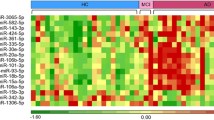
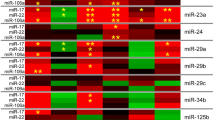
Alzheimer’s disease (AD) is the most common neurodegenerative disease in the older adults. Although much effort has been made in the analyses of diagnostic biomarkers, such as amyloid-β, tau, and neurofilament light chain, identifying peripheral blood-based biomarkers is in extremely urgent need for their minimal invasiveness and more convenience. Here we characterized the miRNA profile by RNA sequencing in human serum exosomes from AD patients and healthy controls (HC) to investigate its potential for AD diagnosis. Subsequently, Gene Ontology analysis and pathway analysis were performed for the targeted genes from the differentially expressed miRNAs. These basic functions were differentially enriched, including cell adhesion, regulation of transcription, and the ubiquitin system. Functional network analysis highlighted the pathways of proteoglycans in cancer, viral carcinogenesis, signaling pathways regulating pluripotency of stem cells, and cellular senescence in AD. A total of 24 miRNAs showed significantly differential expression between AD and HC with more than ± 2.0-fold change at p value < 0.05 and at least 50 reads for each sample. Logistic regression analysis established a model for AD prediction by serum exosomal miR-30b-5p, miR-22-3p, and miR-378a-3p. Sequencing results were validated using quantitative reverse transcription PCR. The data showed that miR-30b-5p, miR-22-3p, and miR-378a-3p were significantly deregulated in AD, with area under the curve (AUC) of 0.668, 0.637, and 0.718, respectively. The combination of the three miRs gained a better diagnostic capability with AUC of 0.880. This finding revealed a miR panel as potential biomarker in the peripheral blood to distinguish AD from HC.





Similar content being viewed by others

Data Availability
All datasets generated or analyzed during the study are included in this published article and its supplementary information files.
References
Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC, Marcus DS, Cairns NJ et al (2012) Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med 367:795–804
Jack CR Jr, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, Shaw LM, Vemuri P et al (2013) Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 12:207–216
Oxtoby NP, Young AL, Cash DM, Benzinger TLS, Fagan AM, Morris JC, Bateman RJ, Fox NC et al (2018) Data-driven models of dominantly-inherited Alzheimer’s disease progression. Brain 141:1529–1544
Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, Holtzman DM, Jagust W et al (2018) NIA-AA Research Framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement 14:535–562
Schindler SE, Li Y, Todd KW, Herries EM, Henson RL, Gray JD, Wang G, Graham DL et al (2019) Emerging cerebrospinal fluid biomarkers in autosomal dominant Alzheimer’s disease. Alzheimers Dement 15:655–665
Bos I, Vos S, Verhey F, Scheltens P, Teunissen C, Engelborghs S, Sleegers K, Frisoni G et al (2019) Cerebrospinal fluid biomarkers of neurodegeneration, synaptic integrity, and astroglial activation across the clinical Alzheimer’s disease spectrum. Alzheimers Dement 15:644–654
Gordon BA, Blazey TM, Su Y, Hari-Raj A, Dincer A, Flores S, Christensen J, McDade E et al (2018) Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer’s disease: a longitudinal study. Lancet Neurol 17:241–250
Rabinovici GD, Gatsonis C, Apgar C, Chaudhary K, Gareen I, Hanna L, Hendrix J, Hillner BE et al (2019) Association of amyloid positron emission tomography with subsequent change in clinical management among medicare beneficiaries with mild cognitive impairment or dementia. JAMA 321:1286–1294
Preische O, Schultz SA, Apel A, Kuhle J, Kaeser SA, Barro C, Graber S, Kuder-Buletta E et al (2019) Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nat Med 25:277–283
Park JC, Han SH, Yi D, Byun MS, Lee JH, Jang S, Ko K, Jeon SY et al (2019) Plasma tau/amyloid-beta1-42 ratio predicts brain tau deposition and neurodegeneration in Alzheimer’s disease. Brain 142:771–786
Nakamura A, Kaneko N, Villemagne VL, Kato T, Doecke J, Dore V, Fowler C, Li QX et al (2018) High performance plasma amyloid-beta biomarkers for Alzheimer’s disease. Nature 554:249–254
Mielke MM, Hagen CE, Xu J, Chai X, Vemuri P, Lowe VJ, Airey DC, Knopman DS et al (2018) Plasma phospho-tau181 increases with Alzheimer’s disease clinical severity and is associated with tau- and amyloid-positron emission tomography. Alzheimers Dement 14:989–997
Hampel H, O’Bryant SE, Molinuevo JL, Zetterberg H, Masters CL, Lista S, Kiddle SJ, Batrla R et al (2018) Blood-based biomarkers for Alzheimer disease: mapping the road to the clinic. Nat Rev Neurol 14:639–652
Gebert LFR, MacRae IJ (2019) Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 20:21–37
Bartel DP (2018) Metazoan MicroRNAs. Cell 173:20–51
Dehghani R, Rahmani F, Rezaei N (2018) MicroRNA in Alzheimer’s disease revisited: implications for major neuropathological mechanisms. Rev Neurosci 29:161–182
Millan MJ (2017) Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: an integrative review. Prog Neurobiol 156:1–68
Qian Q, Zhang J, He FP, Bao WX, Zheng TT, Zhou DM, Pan HY, Zhang H et al (2019) Down-regulated expression of microRNA-338-5p contributes to neuropathology in Alzheimer’s disease. FASEB J 33:4404–4417
Song Y, Hu M, Zhang J, Teng ZQ, Chen C (2019) A novel mechanism of synaptic and cognitive impairments mediated via microRNA-30b in Alzheimer’s disease. EBioMedicine 39:409–421
Wang X, Liu D, Huang HZ, Wang ZH, Hou TY, Yang X, Pang P, Wei N et al (2018) A novel MicroRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in Alzheimer’s disease. Biol Psychiatry 83:395–405
Idda ML, Munk R, Abdelmohsen K, Gorospe M (2018) Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip Rev RNA 9:e1463. https://doi.org/10.1002/wrna.1463
Wiedrick JT, Phillips JI, Lusardi TA, McFarland TJ, Lind B, Sandau US, Harrington CA, Lapidus JA et al (2019) Validation of microRNA biomarkers for Alzheimer’s disease in human cerebrospinal fluid. J Alzheimers Dis 67:875–891
Nagaraj S, Zoltowska KM, Laskowska-Kaszub K, Wojda U (2019) microRNA diagnostic panel for Alzheimer’s disease and epigenetic trade-off between neurodegeneration and cancer. Ageing Res Rev 49:125–143
Kumar S, Reddy PH (2018) MicroRNA-455-3p as a potential biomarker for Alzheimer’s disease: an update. Front Aging Neurosci 10:41
Guo R, Fan G, Zhang J, Wu C, Du Y, Ye H, Li Z, Wang L et al (2017) A 9-microRNA signature in serum serves as a noninvasive biomarker in early diagnosis of Alzheimer’s disease. J Alzheimers Dis 60:1365–1377
Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J et al (2019) Reassessment of exosome composition. Cell 177(428-445):e18
van Niel G, D’Angelo G, Raposo G (2018) Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19:213–228
Shah R, Patel T, Freedman JE (2018) Circulating extracellular vesicles in human disease. N Engl J Med 379:958–966
Riancho J, Vazquez-Higuera JL, Pozueta A, Lage C, Kazimierczak M, Bravo M, Calero M, Gonalezalez A et al (2017) MicroRNA profile in patients with Alzheimer’s disease: analysis of miR-9-5p and miR-598 in raw and exosome enriched cerebrospinal fluid samples. J Alzheimers Dis 57:483–491
Gui Y, Liu H, Zhang L, Lv W, Hu X (2015) Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 6:37043–37053
Yang TT, Liu CG, Gao SC, Zhang Y, Wang PC (2018) The serum exosome derived microRNA-135a, -193b, and -384 were potential Alzheimer’s disease biomarkers. Biomed Environ Sci 31:87–96
Liu CG, Song J, Zhang YQ, Wang PC (2014) MicroRNA-193b is a regulator of amyloid precursor protein in the blood and cerebrospinal fluid derived exosomal microRNA-193b is a biomarker of Alzheimer’s disease. Mol Med Rep 10:2395–2400
Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, Smalheiser NR (2015) Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS One 10:e0139233
Cheng L, Doecke JD, Sharples RA, Villemagne VL, Fowler CJ, Rembach A, Martins RN, Rowe CC et al (2015) Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry 20:1188–1196
McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr, Kawas CH, Klunk WE, Koroshetz WJ et al (2011) The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7:263–269
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128
Dweep H, Gretz N (2015) miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 12:697
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Cheng L, Sharples RA, Scicluna BJ, Hill AF (2014) Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles 26:eCollection
Moradifard S, Hoseinbeyki M, Ganji SM, Minuchehr Z (2018) Analysis of microRNA and gene expression profiles in Alzheimer’s disease: a meta-analysis approach. Sci Rep 8:4767
Liu G, Jiang Y, Wang P, Feng R, Jiang N, Chen X, Song H, Chen Z (2012) Cell adhesion molecules contribute to Alzheimer’s disease: multiple pathway analyses of two genome-wide association studies. J Neurochem 120:190–198
Ahmad K, Baig MH, Mushtaq G, Kamal MA, Greig NH, Choi I (2017) Commonalities in biological pathways, genetics, and cellular mechanism between Alzheimer disease and other neurodegenerative diseases: an in silico-updated overview. Curr Alzheimer Res 14:1190–1197
Mendes-Silva AP, Pereira KS, Tolentino-Araujo GT, Nicolau Ede S, Silva-Ferreira CM, Teixeira AL, Diniz BS (2016) Shared biologic pathways between Alzheimer disease and major depression: a systematic review of microRNA expression studies. Am J Geriatr Psychiatry 24:903–912
Tramutola A, Triani F, Di Domenico F, Barone E, Cai J, Klein JB, Perluigi M, Butterfield DA (2018) Poly-ubiquitin profile in Alzheimer disease brain. Neurobiol Dis 118:129–141
Tramutola A, Di Domenico F, Barone E, Perluigi M, Butterfield DA (2016) It is all about (U)biquitin: role of altered ubiquitin-proteasome system and UCHL1 in Alzheimer disease. Oxidative Med Cell Longev 2016:2756068
Tai HC, Serrano-Pozo A, Hashimoto T, Frosch MP, Spires-Jones TL, Hyman BT (2012) The synaptic accumulation of hyperphosphorylated tau oligomers in Alzheimer disease is associated with dysfunction of the ubiquitin-proteasome system. Am J Pathol 181:1426–1435
Tan SH, Karri V, Tay NWR, Chang KH, Ah HY, Ng PQ, Ho HS, Keh HW et al (2019) Emerging pathways to neurodegeneration: dissecting the critical molecular mechanisms in Alzheimer’s disease, Parkinson’s disease. Biomed Pharmacother 111:765–777
Barrera-Ocampo A, Arlt S, Matschke J, Hartmann U, Puig B, Ferrer I, Zurbig P, Glatzel M et al (2016) Amyloid-beta precursor protein modulates the sorting of Testican-1 and contributes to its accumulation in brain tissue and cerebrospinal fluid from patients with Alzheimer disease. J Neuropathol Exp Neurol 75:903–916
Itzhaki RF, Lathe R, Balin BJ, Ball MJ, Bearer EL, Braak H, Bullido MJ, Carter C et al (2016) Microbes and Alzheimer’s disease. J Alzheimers Dis 51:979–984
Wozniak MA, Mee AP, Itzhaki RF (2009) Herpes simplex virus type 1 DNA is located within Alzheimer’s disease amyloid plaques. J Pathol 217:131–138
Waldau B, Shetty AK (2008) Behavior of neural stem cells in the Alzheimer brain. Cell Mol Life Sci 65:2372–2384
Wadhwani AR, Affaneh A, Van Gulden S, Kessler JA (2019) Neuronal apolipoprotein E4 increases cell death and phosphorylated tau release in Alzheimer disease. Ann Neurol 85:726–739
Wei H, Xu Y, Xu W, Zhou Q, Chen Q, Yang M, Feng F, Liu Y et al (2018) Serum exosomal miR-223 serves as a potential diagnostic and prognostic biomarker for dementia. Neuroscience 379:167–176
Li H, Dai S, Zhen T, Shi H, Zhang F, Yang Y, Kang L, Liang Y et al (2014) Clinical and biological significance of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer 50:1207–1221
Pellatt DF, Stevens JR, Wolff RK, Mullany LE, Herrick JS, Samowitz W, Slattery ML (2016) Expression profiles of miRNA subsets distinguish human colorectal carcinoma and normal colonic mucosa. Clin Transl Gastroenterol 7:e152
Megiorni F, Cialfi S, McDowell HP, Felsani A, Camero S, Guffanti A, Pizer B, Clerico A et al (2014) Deep Sequencing the microRNA profile in rhabdomyosarcoma reveals down-regulation of miR-378 family members. BMC Cancer 14:880
Honegger A, Schilling D, Bastian S, Sponagel J, Kuryshev V, Sultmann H, Scheffner M, Hoppe-Seyler K et al (2015) Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog 11:e1004712
Banerjee S, Karunagaran D (2019) An integrated approach for mining precise RNA-based cervical cancer staging biomarkers. Gene 712:143961
Ikeda K, Horie-Inoue K, Ueno T, Suzuki T, Sato W, Shigekawa T, Osaki A, Saeki T et al (2015) miR-378a-3p modulates tamoxifen sensitivity in breast cancer MCF-7 cells through targeting GOLT1A. Sci Rep 5:13170
Arvidsson Y, Rehammar A, Bergstrom A, Andersson E, Altiparmak G, Sward C, Wangberg B, Kristiansson E et al (2018) miRNA profiling of small intestinal neuroendocrine tumors defines novel molecular subtypes and identifies miR-375 as a biomarker of patient survival. Mod Pathol 31:1302–1317
Jahanbani I, Al-Abdallah A, Ali RH, Al-Brahim N, Mojiminiyi O (2018) Discriminatory miRNAs for the management of papillary thyroid carcinoma and noninvasive follicular thyroid neoplasms with papillary-like nuclear features. Thyroid 28:319–327
Velazquez-Torres G, Shoshan E, Ivan C, Huang L, Fuentes-Mattei E, Paret H, Kim SJ, Rodriguez-Aguayo C et al (2018) A-to-I miR-378a-3p editing can prevent melanoma progression via regulation of PARVA expression. Nat Commun 9:461
Wang M, Sun X, Yang Y, Jiao W (2018) Long non-coding RNA OIP5-AS1 promotes proliferation of lung cancer cells and leads to poor prognosis by targeting miR-378a-3p. Thorac Cancer 9:939–949
Wang Y, Zhang X, Wang Z, Hu Q, Wu J, Li Y, Ren X, Wu T et al (2018) LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett 434:172–183
Xu ZH, Yao TZ, Liu W (2018) miR-378a-3p sensitizes ovarian cancer cells to cisplatin through targeting MAPK1/GRB2. Biomed Pharmacother 107:1410–1417
Zhu P, Ge N, Liu D, Yang F, Zhang K, Guo J, Liu X, Wang S et al (2018) Preliminary investigation of the function of hsa_circ_0006215 in pancreatic cancer. Oncol Lett 16:603–611
Chen CL, Lin CH, Li AL, Huang CC, Shen BY, Chiang YR, Fang PL, Chang HC et al (2019) Plasma miRNA profile is a biomarker associated with urothelial carcinoma in chronic hemodialysis patients. Am J Physiol Ren Physiol 316:F1094–F1102
Fu W, Hong Z, You X, Din J, Chen B, Zhao B, Yuan G, Li Q (2019) Enhancement of anticancer activity of docetaxel by combination with Fuzheng Yiliu decoction in a mouse model of castration-resistant prostate cancer. Biomed Pharmacother 118:109374
Yang J, Zhang Y, Song H (2019) A disparate role of RP11-424C20.2/UHRF1 axis through control of tumor immune escape in liver hepatocellular carcinoma and thymoma. Aging (Albany NY) 11:6422–6439
Cui Y, Xie M, Zhang Z (2020) LINC00958 involves in bladder cancer through sponging miR-378a-3p to elevate IGF1R. Cancer Biother Radiopharm 35:776–788. https://doi.org/10.1089/cbr.2019.3300
Zhang C, Wu S (2020) microRNA -378a-3p restrains the proliferation of retinoblastoma cells but promotes apoptosis of retinoblastoma cells via inhibition of FOXG1. Invest Ophthalmol Vis Sci 61:31
Greco S, Perfetti A, Fasanaro P, Cardani R, Capogrossi MC, Meola G, Martelli F (2012) Deregulated microRNAs in myotonic dystrophy type 2. PLoS One 7:e39732
Siracusa J, Koulmann N, Bourdon S, Goriot ME, Banzet S (2016) Circulating miRNAs as biomarkers of acute muscle damage in rats. Am J Pathol 186:1313–1327
Wei X, Li H, Yang J, Hao D, Dong D, Huang Y, Lan X, Plath M et al (2017) Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis 8:e3153
Wei X, Li H, Zhang B, Li C, Dong D, Lan X, Huang Y, Bai Y et al (2016) miR-378a-3p promotes differentiation and inhibits proliferation of myoblasts by targeting HDAC4 in skeletal muscle development. RNA Biol 13:1300–1309
Florian A, Patrascu A, Tremmel R, Rosch S, Sechtem U, Schwab M, Schaeffeler E, Yilmaz A (2018) Identification of cardiomyopathy-associated circulating miRNA biomarkers in muscular dystrophy female carriers using a complementary cardiac imaging and plasma profiling approach. Front Physiol 9:1770
Huang N, Wang J, Xie W, Lyu Q, Wu J, He J, Qiu W, Xu N et al (2015) MiR-378a-3p enhances adipogenesis by targeting mitogen-activated protein kinase 1. Biochem Biophys Res Commun 457:37–42
Chen W, Zhao W, Yang A, Xu A, Wang H, Cong M, Liu T, Wang P et al (2017) Integrated analysis of microRNA and gene expression profiles reveals a functional regulatory module associated with liver fibrosis. Gene 636:87–95
Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, Ha CS, Kim SW et al (2016) MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun 7:10993
Caserta S, Mengozzi M, Kern F, Newbury SF, Ghezzi P, Llewelyn MJ (2017) Severity of systemic inflammatory response syndrome affects the blood levels of circulating inflammatory-relevant microRNAs. Front Immunol 8:1977
Dubois-Camacho K, Diaz-Jimenez D, De la Fuente M, Quera R, Simian D, Martinez M, Landskron G, Olivares-Morales M et al (2019) Inhibition of miR-378a-3p by inflammation enhances IL-33 levels: a novel mechanism of alarmin modulation in ulcerative colitis. Front Immunol 10:2449
Sud N, Zhang H, Pan K, Cheng X, Cui J, Su Q (2017) Aberrant expression of microRNA induced by high-fructose diet: implications in the pathogenesis of hyperlipidemia and hepatic insulin resistance. J Nutr Biochem 43:125–131
Zhang T, Shi H, Liu N, Tian J, Zhao X, Steer CJ, Han Q, Song G (2020) Activation of microRNA-378a-3p biogenesis promotes hepatic secretion of VLDL and hyperlipidemia by modulating ApoB100-Sortilin1 axis. Theranostics 10:3952–3966
Santangelo L, Bordoni V, Montaldo C, Cimini E, Zingoni A, Battistelli C, D’Offizi G, Capobianchi MR et al (2018) Hepatitis C virus direct-acting antivirals therapy impacts on extracellular vesicles microRNAs content and on their immunomodulating properties. Liver Int 38:1741–1750
Wang-Renault SF, Boudaoud S, Nocturne G, Roche E, Sigrist N, Daviaud C, Bugge Tinggaard A, Renault V et al (2018) Deregulation of microRNA expression in purified T and B lymphocytes from patients with primary Sjogren’s syndrome. Ann Rheum Dis 77:133–140
Muller-Deile J, Dannenberg J, Schroder P, Lin MH, Miner JH, Chen R, Brasen JH, Thum T et al (2017) Podocytes regulate the glomerular basement membrane protein nephronectin by means of miR-378a-3p in glomerular diseases. Kidney Int 92:836–849
Chen T, Wang C, Yu H, Ding M, Zhang C, Lu X, Zhang CY (2019) Increased urinary exosomal microRNAs in children with idiopathic nephrotic syndrome. EBioMedicine 39:552–561
Krist B, Florczyk U, Pietraszek-Gremplewicz K, Jozkowicz A, Dulak J (2015) The role of miR-378a in metabolism, angiogenesis, and muscle biology. Int J Endocrinol 2015:281756
Machado IF, Teodoro JS, Palmeira CM, Rolo AP (2020) miR-378a: a new emerging microRNA in metabolism. Cell Mol Life Sci 77:1947–1958
Ramanan VK, Saykin AJ (2013) Pathways to neurodegeneration: mechanistic insights from GWAS in Alzheimer’s disease, Parkinson’s disease, and related disorders. Am J Neurodegener Dis 2:145–175
Ji Q, Wang X, Cai J, Du X, Sun H, Zhang N (2019) MiR-22-3p regulates amyloid beta deposit in mice model of Alzheimer’s disease by targeting mitogen-activated protein kinase 14. Curr Neurovasc Res 16(5):473–480
Brennan S, Keon M, Liu B, Su Z, Saksena NK (2019) Panoramic visualization of circulating microRNAs across neurodegenerative diseases in humans. Mol Neurobiol 56(11):7380–7407
Acknowledgements
The authors would like to thank all donors and their families for the blood samples provided in the study. Without their selfless contribution, the research could not have proceeded. Furthermore, the authors thank Dr. Shan Yan for his thoughtful discussion of the manuscript.
Funding
This work was supported by Science Foundation of Shanghai Municipal Commission of Science and Technology (19ZR1439300) and Project of Shanghai Municipal Health Commission (201640131).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
This study was performed in line with the principles of the Declaration of Helsinki. Approval was obtained from the ethics committee of Jinshan Branch of Shanghai Sixth People’s Hospital. Informed consent was obtained from all individual participants and the legally authorized representatives of the AD patients included in the study.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Fig. 1
Mapping rate of the NGS generated reads mapped to the reference sequences for each sample. Prefix “A” indicates AD; Prefix “B” indicates HC. (PNG 210 kb)
Supplementary Table 1
All the relevant information of the detected miRNAs, including chromosome, start, end, strand, fold change, and p-value. Overall, 1957 miRNAs were detected in our study. A total of 207 miRNAs were differentially expressed in the AD compared with the HC (p-value < 0.05, and ± 1.2-fold change). (XLSX 570 kb)
Supplementary Table 2
Functional Gene Ontology (GO) analysis of genes that were targeted by these deregulated miRNAs. Biological process (BP), cellular component (CC), and molecular function (MF) terms were ranked by p-value (p-value < 0.05). (XLSX 1759 kb)
Supplementary Table 3
Functional pathway analysis of genes that were targeted by these deregulated miRNAs. The pathways were ranked by p-value (p-value < 0.05). (XLSX 103 kb)
Rights and permissions
About this article
Cite this article
Dong, Z., Gu, H., Guo, Q. et al. Profiling of Serum Exosome MiRNA Reveals the Potential of a MiRNA Panel as Diagnostic Biomarker for Alzheimer’s Disease. Mol Neurobiol 58, 3084–3094 (2021). https://doi.org/10.1007/s12035-021-02323-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02323-y