Abstract
The G protein-coupled GABAB receptors, constituted from GABAB1 and GABAB2 subunits, are important regulators of neuronal excitability by mediating long-lasting inhibition. One factor that determines receptor availability and thereby the strength of inhibition is regulated protein degradation. GABAB receptors are constitutively internalized from the plasma membrane and are either recycled to the cell surface or degraded in lysosomes. Lys-63-linked ubiquitination mediated by the E3 ligase Mind bomb-2 (MIB2) is the signal that sorts GABAB receptors to lysosomes. However, it is unknown how Lys-63-linked ubiquitination and thereby lysosomal degradation of the receptors is regulated. Here, we show that Ca2+/calmodulin-dependent protein kinase II (CaMKII) promotes MIB2-mediated Lys-63-linked ubiquitination of GABAB receptors. We found that inhibition of CaMKII in cultured rat cortical neurons increased cell surface GABAB receptors, whereas overexpression of CaMKIIβ, but not CaMKIIα, decreased receptor levels. This effect was conveyed by Lys-63-linked ubiquitination of GABAB1 at multiple sites mediated by the E3 ligase MIB2. Inactivation of the CaMKII phosphorylation site on GABAB1(Ser-867) strongly reduced Lys-63-linked ubiquitination of GABAB receptors and increased their cell surface expression, whereas the phosphomimetic mutant GABAB1(S867D) exhibited strongly increased Lys-63-linked ubiquitination and reduced cell surface expression. Finally, triggering lysosomal degradation of GABAB receptors by sustained activation of glutamate receptors, a condition occurring in brain ischemia, was accompanied with a massive increase of GABAB1(Ser-867) phosphorylation-dependent Lys-63-linked ubiquitination of GABAB receptors. These findings indicate that CaMKIIβ-dependent Lys-63-linked ubiquitination of GABAB1 at multiple sites controls sorting of GABAB receptors to lysosomes for degradation under physiological and pathological condition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
γ-Aminobutyric acid (GABA), the main inhibitory neurotransmitter in the brain, activates the heterodimeric G protein-coupled GABAB receptors, which are assembled from GABAB1 and GABAB2 subunits. GABAB receptors are abundantly expressed in pre- and postsynaptic compartments of inhibitory as well as excitatory neurons to regulate their excitability [1]. GABAB receptors are expressed by the majority of neurons and are involved in the regulation of virtually all important brain functions, such as neuronal network activity, synaptic plasticity, and neuronal development [2,3,4,5]. Accordingly, dysfunction of GABAB receptor signaling has been implicated in a variety of neurological disorders [6,7,8,9].
For understanding the contribution of GABAB receptors to physiological and pathological mechanisms, the elucidation of its regulation is essential. A main factor regulating GABAB receptor signaling is the dynamic control of their cell surface expression. In this respect, protein degradation is one important mechanism that regulates receptor availability. The two main cellular protein degradation systems, which are proteasomes and lysosomes, contribute to the regulation of cell surface GABAB receptors in different cellular compartments. In the endoplasmic reticulum (ER), the amount of newly synthesized GABAB receptors that are trafficked to the cell surface is determined by proteasomal degradation via the ER-associated degradation (ERAD) machinery [10]. The activity state of the neuron controls the rate of Lys-48-linked ubiquitination of the GABAB2 subunit required for proteasomal receptor degradation [11]. In contrast, GABAB receptors internalized from the cell surface are degraded in lysosomes [12,13,14,15,16]. Sorting the receptors to lysosomes is mediated most likely via the endosomal sorting complex required for transport (ESCRT) machinery [16], which targets ubiquitinated membrane proteins to lysosomes. Accordingly, lysosomal degradation of GABAB receptors depends on Lys-63-linked ubiquitination of the GABAB1 subunit at multiple sites mediated by the E3 ubiquitin ligase MIB2 [17]. However, the mechanism that regulates Lys-63-linked ubiquitination of GABAB1, and thereby lysosomal degradation of the receptors, was unknown. Here, we show that phosphorylation of Ser-867 of GABAB1 by CaMKIIβ regulates the extent of MIB2-mediated K63-linked ubiquitination of GABAB1 and thereby the amount of lysosomal degradation.
Materials and Methods
Antibodies
Mouse anti-HA (clone HA-7, 1:1000 for immunofluorescence, 1:500 for in situ PLA, Sigma-Aldrich catalog no. H9658, lot no. 024M4773), rabbit CaMKII (1:500 for immunofluorescence, 1:100 for in situ PLA; Abcam catalog no. ab52476, lot no. GR181543-20), mouse EEA1 (1:100 for in situ PLA; BD Biosciences catalog no. 61047, lot no. 76250), rabbit GABAB1b directed against the N-terminus of GABAB1b (affinity-purified, 1:200 for immunofluorescence, custom made by GenScript) [18], rabbit GABAB2 directed against the N-terminus of GABAB2 (affinity-purified, 1:500 for immunofluorescence; custom made by GenScript) [19], guinea pig GABAB2 (1:500 for immunofluorescence; Millipore catalog no. AB2255, lot no. 2484228), mouse GABAB1 (1:50 for in situ PLA; NeuroMab, clone N93A/49, catalog no. 75-183), rabbit ubiquitin K48-specific (clone Apu2, 1:50 for in situ PLA; Millipore, catalog no. 05-1307, lot no. 2385989), rabbit ubiquitin K63-specific (clone Apu3, 1:50 for in situ PLA; Millipore, catalog no. 05-1308, lot no. 2575910), rabbit MIB2 (1: 1000 for immunofluorescence, 1:250 for in situ PLA; MyBioSource, catalog no. MBS2014413, lot no. A20160407515), mouse phosphoserine (PSR-45, 1:150 for in situ PLA; Sigma-Aldrich, catalog no. P5747, lot no. 014M4791V), goat Rab7 (1:250 for in situ PLA; Santa Cruz Biotechnology, catalog no. sc-11303, lot no. BO911), and mouse Rab11 (clone 47, 1:25 for in situ PLA; Millipore, catalog no. 05-853, lot no. JBC1868959). Secondary antibodies were labeled with either Alexa Fluor 488 (1:1000, Invitrogen), Cy-3 (1:500, Jackson ImmunoResearch Laboratories), or Cy-5 (1:300, Jackson ImmunoResearch Laboratories).
Drugs
The following chemicals were used for this study: glutamate (50 μM, Sigma-Aldrich) and KN93 (10 μM, Tocris Bioscience). [3H]CGP 54626 (30 Ci/mmol) was purchased from ANAWA Trading SA and CGP 56999A was kindly provided by Novartis.
Plasmids
The following plasmids were used for this study: HA-tagged GABAB1a [20]; GABAB2 [21]; HA-tagged GABAB1a(K689/699R), GABAB1a(K893R), and GABAB1a(K961R) [17]; GFP-tagged CaMKIIα (Addgene plasmid 21226) [22] and its functionally inactive mutant GFP-tagged CaMKIIα(K42M) (here designated CaMKIIα(DN)) (kindly provided by U. Bayer, University of Colorado Denver-AMC); GFP-tagged CaMKIIβ (Addgene plasmid 21227) [22] and its functionally inactive mutant EGFP-tagged CaMKIIβ(K43R) (here designated CaMKIIβ(DN)) (Addgene plasmid 21225) [23]; HA-tagged ubiquitin (Addgene plasmid 17608), HA-tagged ubiquitin (KO) (Addgene plasmid 17603), HA-tagged ubiquitin (K63) (Addgene plasmid 17606), and HA-tagged ubiquitin (K48R) (Addgene plasmid 17604) [24]; and HA-tagged ubiquitin (K63R) (kindly provided by L.-Y. Liu-Chen, Temple University, Philadelphia, USA).
Mutation of GABAB1a
Serine 867 in GABAB1a was mutated either to alanine or to aspartate by GenScript.
Culture and Transfection of Cortical Neurons
Primary cultures (co-cultures of neurons and glia cells) of the cerebral cortex were prepared from 18-day-old embryos of Wistar rats as described previously [25]. Neurons were used after 11 to 15 days in culture. Plasmid DNA was transfected into neurons by magnetofection using Lipofectamine 2000 (Invitrogen) and CombiMag (OZ Biosciences) exactly as specified in Buerli et al. [25]. In our hands, this method reliably yields 50–100 transfected neurons, when transfection was performed with 11–13-day-old cultures.
Culture and Transfection of HEK 293 Cells
HEK (human embryonic kidney) 293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco Life Technologies) containing 10% fetal bovine serum (Gibco Life Technologies) and penicillin/streptomycin (Gibco Life Technologies). Plasmids were introduced into HEK 293 cells using the polyethylenimine method according to the jet-PEI protocol (Polyplus Transfection).
Immunocytochemistry and Confocal Laser Scanning Microscopy
Immunofluorescence staining was done as described previously [13, 26]. For detection of total GABAB receptors, neurons were fixed for 15–20 min at room temperature with 4% paraformaldehyde followed by permeabilization with 0.2% Triton X-100 and immunostaining. For analysis of cell surface GABAB receptors, living neurons were incubated with antibodies recognizing the extracellularly located N-terminal domain of GABAB1 or GABAB2 for 1 h at 4 °C. After washing, the neurons were incubated with fluorophore-labeled secondary antibodies for 1 h at 4 °C and subsequently fixed with 4% paraformaldehyde.
Images of stained neurons were recorded by laser scanning confocal microscopy (LSM 510 Meta, LSM 700 or LSM 710; Zeiss). Five to eight optical sections spaced by 0.3 μm were taken with a ×40, ×63, or ×100 plan-fluar oil differential interference contrast objective (Zeiss) at a resolution of 1024 × 1024 pixels. Total and cell surface fluorescence signals were quantified using the ImageJ software as described previously [26]. To determine cell surface expression of GABAB receptors, all optical sections were merged into one image. Then, the inner as well outer border of somatic staining was carefully outlined and the mean intensity values were measured. Values for cell surface signals were obtained by subtracting the values of the inner border from those of the outer border. For analysis of total GABAB receptor expression, the somata of neurons were carefully outlined and the mean intensity values were measured. For each image, background staining was determined in an area containing no specific signal and subtracted from the mean intensity values. All values were normalized to the analyzed cell area.
Proteins overexpressed in neurons were either tagged with GFP or the HA epitope, which were used to identify transfected neurons and to judge the protein expression level. In case of overexpression studies, transfected neurons with comparable expression of the protein of interested were included into the analysis.
In Situ Proximity Ligation Assay
In situ proximity ligation assay (PLA) is an antibody-based technology for the detection of protein-protein interactions and posttranslational modifications of proteins [27, 28]. We applied in situ PLA for the analysis of GABAB receptor K48-linked or K63-linked ubiquitination, serine phosphorylation, and CaMKII/GABAB receptor interaction as well as for the co-localization with the endosomal marker proteins EEA1, Rab7, and Rab11. In situ PLA was performed using Duolink PLA probes and detection reagents (Sigma-Aldrich) according to the manufacturer’s instructions as described previously [26]. Quantification was done by counting individual in situ PLA spots using the Image J software. The optical sections of each image stack were merged into one image and the number of maxima was determined after setting an appropriate noise tolerance (noise tolerance was kept constant for all images of an experiment). The number of spots was normalized to the area analyzed and to the expression level of GABAB receptors. In some experiments, signals were too abundant to resolve individual spots precisely. In those cases, the fluorescence intensity of signals was determined.
Internalization Assay
Neurons were placed on ice and cell surface GABAB receptors were labeled with GABAB2 antibodies for 1 h at 4 °C. After extensive washes, neurons were incubated in 37 °C warm medium for exactly 1, 2, 3, 5, or 10 min to permit receptor endocytosis. Endocytosis was terminated by replacing the medium with ice-cold buffer. After incubation with secondary antibody for 1 h at 4 °C, the neurons were fixed with 4% PFA and remaining cell surface GABAB receptors were determined by laser scanning confocal microscopy.
Western Blot Analysis
For Western blot analysis, neuron/glia co-cultures were grown for 13–14 days on 6-cm culture dishes plated with 450,000 cells obtained from the cerebral cortex of E18 rat embryos. After incubation with KN93, cultures were immediately washed twice with ice-cold PBS, harvested, and homogenized by sonication. After protein determination using the Bradford protein assay (BioRad), the samples were incubated with Laemmli sample buffer (BioRad) for 2 h at 37 °C and aliquots containing 30 μg protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5% mini-gels (Mini Protean 3, BioRad). Proteins were transferred onto nitrocellulose membranes in a Mini Trans-Blot Module (BioRad) at 365 mA for 120 min using 192 mM glycine, 25 mM Tris, 0.1% SDS, 20% methanol as transfer buffer. After blotting, the transferred proteins were stained with Amidoblack and immediately imaged using the E-box VX2 gel imager (Vilber). For immunodetection, the blots were blocked for 1–2 h in PBST (PBS pH 7.4, 0.05% Tween 20) containing 5% nonfat dry milk at room temperature, followed by incubation with antisera overnight at 4 °C in PBST containing 5% non-fat dry milk. The blots were then washed five times for 10 min with TBST and incubated with secondary antibodies conjugated to horseradish peroxidase for 1 h at room temperature. Following extensive washing (see above), immunoreactivity was detected by the chemoluminescence method (SuperSignal West Dura, Thermo Scientific) using a Fujifilm LAS-1000 imager. Immunoreactivity was quantified with the Image Studio software (LI-CORE Biosciences) and normalized to total protein in the corresponding lanes (determined by Amidoblack staining, see above).
Radioligand Binding
For radioligand binding, 450,000 cells derived from the cerebral cortex of E18 rat embryos were plated onto 6-cm polylysine-coated culture dishes and kept in culture for 14 days. After drug incubation, cells were immediately washed two times with ice-cold PBS and harvested. Cells were resuspended in 500 μl binding buffer containing protease inhibitors (50 mM Tris pH 7.4, 2.5 mM CaCl2, complete mini, Sigma-Aldrich) and homogenized using a Potter S homogenizer (B. Braun Biotech International). Aliquots of the homogenate were incubated with 6.7 nM [3H]CGP 54626 in binding buffer for 90 min at room temperature. The incubation was terminated by rapid filtration onto glass fiber filters using a 12-channnel semi-automated cell harvester (Scatron) and washed with ice-cold binding buffer. Non-specific [3H]CGP 54626 binding was determined in the presence of 10 μM CGP 56999A. The radioactivity retained by the filters was determined by liquid scintillation counting using a Tricarb 2500 liquid scintillation analyzer.
Statistics
The statistical analyses were done with GraphPad Prism 5. The tests used and p values are given in the figure legends. Differences were considered statistically significant when p < 0.05.
Results
CaMKIIβ Controls Expression of GABAB Receptors
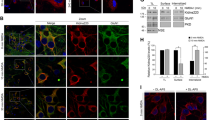
Sustained activation of glutamate receptors, which occurs in cerebral ischemia, downregulates GABAB receptors by preferentially sorting constitutively internalized receptors to lysosomes for degradation instead of recycling them to the cell surface [29,30,31,32]. Phosphorylation of GABAB1 at Ser-867 by CaMKII plays a key role in this mechanism [29]. However, it was unknown whether CaMKII is also involved in the regulation of GABAB receptor expression under normal physiological conditions. Therefore, we tested whether inhibition of CaMKII by KN93 affects total expression of GABAB receptors in cultured cortical neurons. Incubation of neurons with KN93 transiently increased the expression of GABAB1 and GABAB2, which peaked at about 7.5 min as tested by Western blotting (Fig. 1a). This was confirmed by radioligand binding experiments on homogenates prepared from the cultures using the GABAB receptor antagonist [3H]CGP 54626 (Fig. 1b). Finally, we monitored cell surface expression of GABAB receptors by staining living neurons at 4 °C with antibodies directed against the extracellular located N-terminal domain of the receptors. In line with the experiments on total receptor expression, incubation of neurons with KN93 for 7.5 min considerably increased the level of GABAB receptors at the cell surface (148 ± 64% of control; Fig. 1c). The cause for the transient nature of blocking CaMKII on cell surface expression of the receptors is currently unclear. It might well be that a so far unknown homeostatic response of the neurons downregulates the increased cell surface receptors to normal levels. However, our results clearly indicate that the expression of the receptors in the plasma membrane is indeed controlled by CaMKII under basal conditions.
CaMKII regulates expression of GABAB receptors. Neuronal cultures were incubated for the indicated times with the CaMKII inhibitor KN93 (10 μM) and tested for GABAB receptor expression by Western blotting using antibodies against GABAB1 and GABAB2 (a) or radioligand binding with the GABAB receptor antagonist [3H]CGP 54626 (b). In addition, cell surface expression of GABAB receptors was analyzed by immunocytochemistry using GABAB1 antibodies (c). Untreated cultures served as controls. a Mean ± SD of 8–10 independent cultures for GABAB1 and 7–9 cultures for GABAB2; b Mean ± SD of 4–6 independent cultures; c Mean ± SD of 39–43 neurons from three independent experiments. Scale bar in representative immunofluorescence images depicted in c represents 5 μm. *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA, Dunnett’s multiple comparison test
CaMKIIα and CaMKIIβ are the main CaMKII isoforms expressed in neurons [33]. We therefore tested which isoform is involved in the regulation of GABAB receptors by transfecting neurons with either CaMKIIα, CaMKIIβ, or their functionally inactive mutants (CaMKIIα(DN), CaMKIIβ(DN)) and determined cell surface expression of GABAB receptors using GABAB2 antibodies. While transfecting CaMKIIα or CaMKIIα(DN) did not affect cell surface expression of GABAB receptors, overexpression of CaMKIIβ decreased (43 ± 17% of EGFP-transfected control neurons, Fig. 2a) and its nonfunctional mutant CaMKIIβ(DN) increased cell surface levels of receptors (186 ± 29% of EGFP-transfected control neurons, Fig. 2b).
CaMKIIβ interacts in an activity-dependent manner with GABAB receptors to regulate their cell surface expression. a CaMKIIβ but not CaMKIIα regulates cell surface expression of GABAB receptors. Cortical neurons were transfected with either EGFP (control), CaMKIIα, CaMKIIβ, or their functionally inactive mutants CaMKIIβ(DN) or CaMKIIα(DN). Two days after transfection, cell surface expression of GABAB receptors was assessed using GABAB2 antibodies. Transfected neurons were identified via the GFP-tag of transfected CaMKII constructs (not shown). Left, representative images of the soma of stained neurons (scale bar, 5 μm). Right, the graphs show the quantification of fluorescence intensities. Fluorescence intensities for GABAB2 in EGFP-transfected neurons were set to 100%. The data represent the mean ± SD of 26–30 neurons from three independent experiments. ***p < 0.001, nsp > 0.05; one-way ANOVA, Dunnett’s multiple comparison test. b CaMKIIβ interacts with GABAB receptors in an activity-dependent manner. Neurons were transfected with either EGFP (control), CaMKIIα, or CaMKIIβ and tested for interaction of CaMKII with GABAB receptors in the presence of absence of KN93 by in situ PLA using antibodies directed against CaMKII and GABAB1. Left, representative images of in situ PLA signals (white dots) in the soma of neurons (scale bar, 5 μm). Right, the graph shows the quantification PLA signals. The data represent the mean ± SD of 30 neurons per condition from three independent experiments. ***p < 0.001, nsp > 0.05; one-way ANOVA, Dunnett’s multiple comparison test
It was previously shown that CaMKII interacts with GABAB1 [29]. Next, we tested whether CaMKIIβ or CaMKIIα interacts with GABAB receptors under basal conditions by overexpressing either CaMKIIα or CaMKIIβ in neurons and testing for changes in interaction levels with GABAB receptors using in situ PLA with antibodies against GABAB1 and CaMKII (Fig. 2b). In line with our experiments described above, overexpression of CaMKIIα did not affect the level of GABAB receptor/CaMKII interaction (PLA signal EGFP-transfected control, 100 ± 32%; CaMKIIα, 80 ± 30%), but transfection of CaMKIIβ significantly increased the extent of interaction (216 ± 58% of control). The interaction of CaMKIIβ with GABAB receptors was activity-dependent as blocking CaMKII with KN93 reduced the interaction level in neurons transfected with CaMKIIβ (PLA signal. 110 ± 40% of control).
These results indicate that CaMKIIβ interacts with GABAB receptors in an activity-dependent manner to regulate the cell surface expression of the receptors.
CaMKII Triggers Sorting of GABAB Receptors to Lysosomal Degradation
It had been suggested that phosphorylation of GABAB1 Ser-867 by CaMKII may mediate internalization of the receptors [29]. We therefore tested whether inhibition of CaMKII by KN93 affects the internalization of GABAB receptors. However, blocking CaMKII with KN93 did affect neither the kinetics (half live; control, 1.2 min; KN93, 1.6 min) nor the extent (control, 64 ± 19%; KN93, 65 ± 20%) of receptor internalization (Fig. 3a).
CaMKII triggers sorting of GABAB receptors to lysosomes. a Blocking CaMKII activity does not affect internalization of GABAB receptors. GABAB2 antibody-labeled cell surface receptors of living neurons were incubated for the indicated time intervals at 37 °C to permit endocytosis. Subsequently, remaining cell surface GABAB2-labeled receptors were quantified by immunofluorescence microscopy. The immunofluorescence signals of neurons at time 0 (not exposed to 37 °C) were set to 100%. Data were fitted by nonlinear regression to one phase decay. The data represent the mean ± SD of 35–46 neurons per time point from two independent experiments. b Blocking CaMKII activity decreased the co-localization of GABAB receptors with Rab7 and increased the co-localization with Rab11. Neurons were either treated or not with KN93 and subjected to in situ PLA using antibodies directed against GABAB1 and Rab11 (marker for recycling endosomes), Rab7 (marker for the lysosomal pathway), or EEA1 (marker for early endosomes), respectively. Left, representative images (scale bar 5 μm). Right, quantification of PLA signals (white dots in images). The PLA signals were normalized to the cell area. The control condition was set to 100%. The data represent the mean ± SD of 30 neurons (Rab11) and 40 neurons (Rab7, EEA1) from two independent experiments. ****p < 0.0001, nsp > 0.05; two-tailed unpaired t test
To gain insight into the mechanism that is affected by CaMKII inhibition, we tested the co-localization of GABAB receptors with the endosomal marker proteins EEA1 (marker for early endosomes), Rab7 (marker for late endosomes and lysosomes), and Rab11 (marker for recycling endosomes) [34] using in situ PLA. Inhibition of CaMKII activity in neurons with KN93 did not affect the co-localization of GABAB receptors with EEA1 (94 ± 26% of control; Fig. 3b), supporting our observation of an unchanged internalization rate of the receptors. Instead, blocking CaMKII decreased the co-localization of GABAB receptors with Rab7 (39 ± 28% of control; Fig. 3b) and increased the co-localization of GABAB receptors with Rab11 (145 ± 32% of control; Fig. 3b). Thus, inhibition of CaMKII appears to prevent recruitment of GABAB receptors to the lysosomal degradation pathway (Rab7) and increase recycling of the receptors (Rab11). This observation indicates that basal CaMKII activity is involved in targeting GABAB receptors to lysosomes for degradation.
CaMKIIβ Promotes Lys-63-Linked Ubiquitination of GABAB Receptors
The results so far support the hypothesis that CaMKIIβ regulates lysosomal degradation of GABAB receptors. We had previously shown that lysosomal degradation of GABAB receptors depends on Lys-63-linked ubiquitination of GABAB1 at multiple sites via the E3 ligase MIB2 [17]. It was therefore likely that CaMKIIβ controls Lys-63-linked ubiquitination of GABAB receptors.
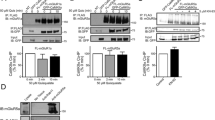
To test this hypothesis, we first analyzed the effect of various ubiquitin mutants on the downregulation of cell surface GABAB receptors induced by overexpression of CaMKIIβ. Overexpression of CaMKIIβ in neurons reduced cell surface GABAB receptors to 43 ± 18% of control neurons transfected with EGFP (Fig. 4). Additional overexpression of wild-type ubiquitin (Ub, 36 ± 25% of control) and a ubiquitin mutant that permits only Lys-63-linked ubiquitination (Ub(K63), 35 ± 18% of control) as well as a mutant that prevents Lys-48-linked ubiquitination (Ub(K48R), 39 ± 24% of control) did not affect CaMKIIβ-induced downregulation of the receptors (43 ± 18% of control). However, overexpressing a ubiquitin mutant in which all lysine residues were mutated to arginine to entirely block polyubiquitination (Ub(KO), 109 ± 22% of control) as well as a ubiquitin mutant that specifically prevents Lys-63-linked ubiquitination (Ub(K63R), 124 ± 40% of control) completely prevented downregulation of the receptors or even increased cell surface expression of the receptors (Fig. 4). These results indicate that downregulation of GABAB receptors induced by overexpression of CaMKIIβ is mediated by Lys-63-linked ubiquitination.
CaMKIIβ-induced downregulation of cell surface GABAB receptors depends on Lys-63-linked ubiquitination. Neurons were transfected with either EGFP (control), CaMKIIβ, or CaMKIIβ plus ubiquitin (Ub) or one of its mutants (Ub(K63): only Lys-63 linkages, Ub(K63R): no Lys-63 linkages, Ub(K48R): no Lys-48 linkages, Ub(KO): no polyubiquitin linkages (only mono-ubiquitination). Two days after transfection, neurons were stained for GABAB receptors using GABAB2 antibodies. Only interference with K63-linked ubiquitination prevented CaMKIIβ-mediated downregulation of the receptors. Representative images of stained neuronal somata (scale bar, 5 μm). The scatter plot shows quantification of fluorescence intensities. The fluorescence signal of neurons transfected with EGFP was set to 100%. The data represent the mean ± SD of 28–30 neurons from two independent experiments. ***p < 0.001, nsp > 0.05; one-way ANOVA, Dunnett’s multiple comparison test
Next, we tested for direct CaMKIIβ-induced Lys-63-linked ubiquitination of GABAB receptors using in situ PLA and antibodies directed against GABAB1 and Lys-63-linked ubiquitin. Inhibition of CaMKII with KN93 significantly reduced basal Lys-63-linked ubiquitination of the receptor (69 ± 43% of control, Fig. 5a). In addition, overexpression of the functionally inactive CaMKIIβ mutant CaMKIIβ(DN) in neurons considerably reduced Lys-63-linked ubiquitination of the receptors (46 ± 24%, Fig. 5b) as compared to neurons transfected with wild-type CaMKIIβ. In contrast, overexpression of the functionally inactive mutant of CaMKIIα (CaMKIIα(DΝ)) had no effect on the level of Lys-63-linked ubiquitinated GABAB receptors as compared to wild-type CaMKIIα. Finally, overexpression of neither CaMKIIα(DN) nor CaMKIIβ(DN) affected Lys-48-linked ubiquitination of the receptors, which is required for their proteasomal degradation (Fig. 5c). These results indicate that CaMKIIβ is involved in the regulation of Lys-63-linked ubiquitination of GABAB receptors.
CaMKII regulates K63-linked ubiquitination of GABAB receptors. a Inhibition of CaMKII activity reduced K63-linked ubiquitination of GABAB receptors. Neurons were incubated for 7.5 min in the absence (control) or presence of KN93 and analyzed for K63-linked ubiquitination of GABAB receptor by in situ PLA using antibodies directed against GABAB1- and K63-linked ubiquitin (white dots in representative images, scale bar 5 μm). Right, quantification of in situ PLA signals. The data represent the mean ± SD of 33 (control) and 30 (KN93) neurons from two independent experiments. **p < 0.01, two-tailed unpaired t test. b, c CaMKIIβ promotes Lys-63-linked ubiquitination of GABAB receptors. Neurons were transfected with either CaMKIIα or CaMKIIβ or their functionally inactive mutants CaMKIIα(DN) or CaMKIIβ(DN). After 2 days, GABAB receptors were tested for Lys-63-linked (b) or Lys-48-linked (c) ubiquitination by in situ PLA using antibodies directed against GABAB1 and Lys-63 or Lys-48 ubiquitin. Left, representative images depicting PLA signals (white dots, scale bars 5 μm). Right, quantification of PLA signals. The data represent the mean ± SD of 17–21 neurons from two independent experiments. **p < 0.01, ****p < 0.0001, nsp > 0.05; two-tailed unpaired t test
We recently identified in GABAB1 several lysine residues (Lys-697/698, Lys-892, and Lys-960) that are substrates for Lys-63-linked ubiquitination and important for targeting the receptors to lysosomal degradation [17]. To further substantiate the finding that CaMKII regulates lysosomal degradation of GABAB receptors by promoting Lys-63-linked ubiquitination of GABAB1, we tested whether blocking CaMKII affects cell surface expression of three GABAB1a(K->R) mutants in which Lys-697/698, Lys-892, or Lys-960 was mutated to arginine (GABAB1a(K697/698R), GABAB1a(K892R), and GABAB1a(K960R)). In contrast to receptors containing wild-type GABAB1, which displayed increased cell surface expression upon treatment with KN93 (GABAB1: 164 ± 149%, GABAB2: 142 ± 91% of control; Fig. 6), the expression level of receptors containing any of the three GABAB1a(K->R) mutants remained unaffected by inhibition of CaMKII (GABAB1a(K697/698R), control: 552 ± 426%, KN93: 490 ± 456%; GABAB1a(K892R), control: 421 ± 284%, KN93: 370 ± 272%; GABAB1a (K960R), control: 273 ± 256%, KN93: 281 ± 270% of wild-type control; GABAB2 co-expressed with GABAB1a (K697/698R), control: 160 ± 115%, KN93: 140 ± 117%; GABAB2 co-expressed with GABAB1a(K892R), control: 184 ± 106%, KN93: 163 ± 88%; GABAB2 co-expressed with GABAB1a(K960R), control: 147 ± 84%, KN93: 145 ± 78% of wild-type control; Fig. 6). In conclusion, these findings suggest that CaMKII is involved in K63-linked ubiquitination of GABAB1 at K697/698 and K892 as well as K960.
Blocking CaMKII did not affect the expression levels of GABAB1a(K->R) mutants. Neurons were transfected with HA-tagged wild-type GABAB1a or HA-tagged GABAB1a (K->R) mutants along with GABAB2 and tested for cell surface expression of GABAB1 and GABAB2 after treating the neurons with 10 μM KN93 for 7.5 min. Left, representative images of untreated neurons (control, left panels) and of neurons treated with KN93 (right panels, scale bar 5 μm). The corresponding graphs show the quantification of fluorescence signals. The fluorescence intensity of neurons transfected with wild-type GABAB1 from untreated neurons (control) was set to 100%. The data represent the mean ± SD of 45–60 neurons per experimental condition derived from three independent experiments. Please note that the considerably lower increase in GABAB2 cell surface expression as compared with mutant GABAB1 was due to the fact that in the case of GABAB1, only transfected subunits were detected (HA-tagged) but in the case of GABAB2, transfected as well as endogenously expressed subunits were detected. ***p < 0.001, nsp > 0.05; two-tailed unpaired t test
CaMKII Regulates Cell Surface Receptors by Promoting MIB2-Mediated Lys-63-Linked Ubiquitination via Phosphorylation of GABAB1 at Ser-867
CaMKII phosphorylates GABAB1 at Ser-867 [29]. To prove that direct phosphorylation of Ser-867 in GABAB1 is involved in the regulation of GABAB receptor cell surface expression, we silenced this phosphorylation site by mutating it to alanine (GABAB1(S867A)) or rendered the site permanently active by mutating it to aspartate (GABAB1(S867D)). First, we verified that CaMKIIβ indeed phosphorylates GABAB1 at Ser-867 by in situ PLA using antibodies directed against GABAB1 and phosphoserine. Upon expression in HEK 293 cells, wild-type GABAB receptors displayed a basal level of serine phosphorylation, which was more than threefold enhanced by co-expression with CaMKIIβ (PLA signal wt-GABAB1: 98 ± 5, GABAB1+CaMKIIβ: 358 ± 14; Fig. 7a). In contrast, GABAB receptors in which Ser-867 of GABAB1 was inactivated by mutation to alanine, only basal level of serine phosphorylation was detected in the presence of CaMKIIβ (PLA signal GABAB1(S867A)+CaMKIIβ: 108 ± 6 compared to wt-GABAB1: 98 ± 5, Fig. 7a). These results confirm that CaMKIIβ specifically phosphorylates GABAB1 at Ser-867.
Phosphorylation of GABAB1 Ser-867 controls cell surface expression and Lys-63-linked ubiquitination of GABAB receptors. a CaMKIIβ phosphorylates GABAB1 at Ser-867. HEK293 cells were transfected either with GABAB1/GABAB2 with or without CaMKIIβ or with GABAB1(S867A)/GABAB2 and CaMKIIβ. Two days after transfection, the cells were tested for serine phosphorylation by in situ PLA using antibodies directed against GABAB1 and phosphoserine. Left, representative images (scale bar, 10 μm; wt, wild type). Right, quantification of PLA signals (white dots in images). The PLA signals were normalized to the cell area as well as to the expression level of GABAB1. The data represent the mean ± SD of 50 cells from two independent experiments. ***p < 0.001, nsp > 0.05; one-way ANOVA, Dunnett’s multiple comparison test. b, c Inactivation or constitutively mimicking phosphorylation of GABAB1 Ser-867 affects cell surface expression (b) as well as Lys-63-linked ubiquitination (c) of the receptors. Neurons were transfected with wild-type HA-tagged GABAB1a (control), with the phosphorylation-deficient HA-tagged GABAB1a(S867A) mutant or with the phosphomimetic HA-tagged GABAB1a(S867D) mutant along with GABAB2 and tested for cell surface expression using HA antibodies (b) as well as for Lys63-linked ubiquitination by in situ PLA with HA and Lys-63 antibodies (c). Left, representative images (scale bars, 5 μm). Right, quantification of fluorescence intensities (b) and PLA signals (c). The data represent the mean ± SD of 23–27 (b) and 18–20 (c) neurons from two independent experiments. *p < 0.05, ***p < 0.001; one-way ANOVA, Dunnett’s multiple comparison test
Next, we tested the effect of inactivating or permanently mimicking phosphorylation of GABAB1 Ser-867 on cell surface expression of the receptors by expressing GABAB1(S867A) or GABAB1(S867D) in cortical neurons. Inactivation of GABAB1 Ser-867 considerably increased cell surface expression of receptors containing GABAB1(S867A) (271 ± 119% of control, Fig. 7b) upon transfection in neurons, whereas receptors containing the phosphomimetic GABAB1(S867D) mutant were expressed at reduced levels (50 ± 22% of control, Fig. 7b). In line with this finding, receptors containing the CaMKII phosphorylation-deficient GABAB1(S867A) mutant exhibited reduced K63-linked ubiquitination (47 ± 32% of control, Fig. 7c), while receptors containing the phosphomimetic GABAB1(S867D) mutant showed a considerably higher level of Lys-63-linked ubiquitination (226 ± 76% of control, Fig. 7c). These findings suggest that CaMKII promotes Lys-63-linked ubiquitination of GABAB receptors via phosphorylation of serine 867 in GABAB1.
We previously showed that Lys-63-linked ubiquitination of GABAB1 is mediated by the ubiquitin E3 ligase MIB2 [17]. We therefore tested for the involvement of MIB2 in CaMKIIβ-induced downregulation of GABAB receptors. Overexpression of MIB2 or CaMKIIβ reduced cell surface expression of GABAB receptors in neurons to a similar extent (MIB2: 40 ± 16%, CaMKIIβ: 45 ± 17% of EGFP-transfected control neurons; Fig. 8a). Co-transfection of MIB2 and CaMKIIβ in neurons did not further increase downregulation of the receptors (43 ± 14% of EGFP-transfected control neurons; Fig. 8a), indicating that both enzymes affect GABAB receptors via the same pathway. In line with this observation, MIB2 was unable to downregulate cell surface GABAB receptors when the nonfunctional CaMKIIβ mutant CaMKIIβ(DN) was simultaneously overexpressed with MIB2 (CaMKIIβ(DN): 195 ± 42%, CaMKIIβ(DN)+MIB2: 200 ± 43% of EGFP-transfected control neurons, Fig. 8a).
The ubiquitin E3 ligase MIB2 is involved in CaMKIIβ-mediated regulation of cell surface GABAB receptors. a Overexpression of MIB2 in addition to CaMKII does not enhance downregulation of cell surface GABAB receptors. Neurons were transfected with the indicated plasmid and tested for cell surface expression of GABAB receptors using GABAB2 antibodies 2 days after transfection. Co-expression of MIB2 did not further enhance downregulation of cell surface receptors, indicating the involvement in the same pathway. Upper panels show representative images (scale bars, 5 μm). The graphs depict quantification of the fluorescence signals. The data represent the mean ± SD of 30 neurons per condition derived from two independent experiments. nsp > 0.05, ****p < 0.0001; two-way ANOVA with Bonferroni’s multiple comparison test (interaction: F(2,234) = 32.78, p < 0.0001). b MIB2-induced downregulation of cell surface receptors depends on phosphorylation of GABAB1 Ser-867. Neurons were transfected with wild-type HA-tagged GABAB1a (control), with the phosphorylation-deficient HA-tagged GABAB1a(S867A) mutant or with the phosphomimetic HA-tagged GABAB1a(S867D) mutant along with GABAB2 and with or without MIB2. Two days after transfection, neurons were tested for cell surface expression of tagged GABAB1 using HA antibodies. Upper panels show representative images (scale bars, 5 μm). The graph depicts quantification of the fluorescence signals. The data represent the mean ± SD of 30 neurons per condition derived from two independent experiments. nsp > 0.05, ****p < 0.0001; two-way ANOVA, Bonferroni’s multiple comparison test (interaction: F(2,174) = 49.09, p < 0.0001). c GABAB1 phospho-mutants display altered MIB2 interaction. HEK 293 cells were transfected with wild-type GABAB1, GABAB1(S867A), or GABAB1(S867D) together with GABAB2 and MIB2. Two days after transfection, cells were analyzed for interaction of GABAB receptors with MIB2 by in situ PLA using antibodies directed against GABAB1 and MIB2. Upper panels show representative images (scale bar, 10 μm). The graph depicts quantification of the PLA signals. The data represent the mean ± SD of 35 neurons per condition derived from two independent experiments. **p < 0.01, ***p < 0.001; one-way ANOVA, Dunnett’s multiple comparison test
Next, we tested whether MIB2-mediated downregulation of GABAB receptors depends on phosphorylation of GABAB1 Ser-867. Co-transfection of GABAB1a,2 and MIB2 in neurons strongly reduced cell surface expression of receptors containing transfected GABAB1a (28 ± 11% of control, Fig. 8b). However, MIB2 failed to affect expression of the elevated or reduced cell surface expression levels of receptors containing GABAB1a(S867A) or GABAB1a(S867D) (S867A: 204 ± 39%, S867A+MIB2: 211 ± 38%, S867D: 23 ± 8%, S867D+MIB2: 24 ± 9% of control, Fig. 8b). Together, these results indicate that phosphorylation of GABAB1 at Ser-867 by CaMKIIβ is required for MIB2-mediated downregulation of GABAB receptors.
Finally, we tested the hypothesis that CaMKII-mediated phosphorylation of GABAB1 might affect the interaction of MIB2 with GABAB receptors. Upon transfection in HEK 293 cells, MIB2 displayed considerably lower interaction with receptors containing the phospho-deficient mutant GABAB1a(S867A) (25 ± 15% of control, Fig. 8c) and a higher level of interaction with receptors containing the phosphomimetic mutant GABAB1a(S867D) (126 ± 44% of control, Fig. 8c) than wild-type receptors. This result indicates that phosphorylation of GABAB1 at Ser-867 promotes the interaction of GABAB receptors with MIB2.
Sustained Activation of Glutamate Receptors Increases Lys-63-Linked Ubiquitination of GABAB Receptors via Phosphorylation of GABAB1 Ser-867
Glutamate-induced downregulation of GABAB receptors via lysosomal degradation depends on CaMKII-mediated phosphorylation of GABAB1 Ser-867 [29] as well as on Lys-63-linked ubiquitination of GABAB1 at Lys-697/698, Lys-892, and Lys-960 [17]. To investigate the role of CaMKII in this process, we first analyzed the effect of sustained glutamate treatment on the expression level of CaMKII as well as on the degree of CaMKII-GABAB receptor interaction. Treatment of neurons for 60 min with glutamate significantly upregulated CaMKII (233 ± 61% of control, Fig. 9a) and considerably increased the interaction of CaMKII with GABAB receptors (165 ± 74% of control, Fig. 9b).
Glutamate-induced downregulation of GABAB receptors. a Prolonged glutamate treatment increased CaMKII expression. Neurons were treated with 50 μM glutamate for 60 min and analyzed after additional 16 h for CaMKII expression. Upper panel shows representative images (scale bar, 5 μm). The data represent the mean ± SD of 45 neurons per experimental condition derived from two independent experiments. ****p < 0.0001, two-tailed unpaired t test. b Prolonged glutamate treatment increased the interaction of CaMKII with GABAB receptors. Neurons were treated with 50 μM glutamate for 60 min and analyzed after additional 16 h for the interaction of CaMKII with GABAB receptors via in situ PLA using CaMKII and GABAB1 antibodies. Upper panel shows representative images (scale bar, 5 μm). The data represent the mean ± SD of 39 neurons per experimental condition derived from two independent experiments. ****p < 0.0001, two-tailed unpaired t test. c, d Glutamate-induced downregulation of GABAB receptors is mediated by GABAB1 Ser-867 phosphorylation-induced Lys-63-linked ubiquitination. Neurons transfected with wild-type HA-tagged GABAB1a, with the phosphorylation-deficient HA-tagged GABAB1a(S867A) mutant or with the phosphomimetic HA-tagged GABAB1a(S867D) mutant along with GABAB2 were incubated for 60 min in the absence (control) or presence of 50 μM glutamate. Neurons were analyzed for cell surface expression of transfected GABAB1 using HA antibodies (c) or analyzed for Lys-63-linked ubiquitination by in situ PLA using HA and Lys-63 antibodies (d). Upper panels show representative images (scale bars, 5 μm). The graphs depict quantification of the fluorescence signals (c) and in situ PLA signals (d). The data represent the mean ± SD of 28–30 (c) and 28–36 (d) neurons per condition derived from three independent experiments. nsp > 0.05, ****p < 0.0001; two-way ANOVA, Bonferroni’s multiple comparison test (c interaction: F(2,170) = 6.27, p < 0.005; d interaction: F(2, 205) = 17.22, p < 0.0001)
We then analyzed whether glutamate-induced K63-linked ubiquitination of GABAB receptors depends on phosphorylation of Ser-867. Wild-type GABAB1 or the two mutants GABAB1(S867A) and GABAB1(S867D) were expressed in cortical neurons and exposed for 60 min to glutamate. Under this condition, cell surface wild-type GABAB receptors were downregulated to 30 ± 20% as compared to untreated controls (Fig. 9c). However, receptors containing the phospho-deficient mutant GABAB1(S867A) showed enhanced cell surface expression under control conditions (240 ± 99% of wild-type controls) and were not affected by glutamate (240 ± 94% of wild-type controls, Fig. 9c). In contrast, receptors containing the phosphomimetic mutant GABAB1(S867D) displayed strongly reduced expression under control conditions (45 ± 24% of wild-type controls, Fig. 9c) and remained unaffected after glutamate treatment (44 ± 23% of wild-type controls, Fig. 9c). A complementary pattern was observed for Lys-63-linked ubiquitination of the respective receptors as tested by in situ PLA. Treatment of cultured neurons with glutamate strongly increased K63-linked ubiquitination of wild-type GABAB receptors (control: 100 ± 60%, glutamate: 247 ± 84%, Fig. 9d), whereas receptors containing the phospho-deficient mutant GABAB1(S867A) displayed only marginal Lys-63-linked ubiquitination under both conditions (control: 10 ± 13%, glutamate: 11 ± 13, Fig. 9d) and the phosphomimetic mutant GABAB1(S867D) exhibited strongly enhanced Lys-63-linked ubiquitination (control: 175 ± 126, glutamate: 181 ± 121, Fig. 9d).
These findings suggest that sustained activation of glutamate receptors induces GABAB1-Ser-867 phosphorylation-mediated K63-linked ubiquitination of GABAB receptors, promoting their lysosomal degradation.
Discussion
The expression of GABAB receptors available for signal transduction at the cell surface critically depends on their rate of degradation. The two main cellular degradation systems—proteasomes and lysosomes—control the abundance of GABAB receptors in distinct cellular compartments in response to the activity state of the neuron [10, 11, 17]. In the ER, the amount of newly synthetized GABAB receptors destined for trafficking to the plasma membrane is regulated by proteasomal degradation via the ERAD machinery [10], whereas cell surface GABAB receptors are degraded in lysosomes [12,13,14,15,16]. Both degradation pathways require ubiquitination of the receptor as targeting signals. Lys-48-linked ubiquitination of GABAB2 at Lys-767/771 tags GABAB receptors for proteasomal degradation [10] and Lys-63-linked ubiquitination of GABAB1 at several sites sorts the receptors to lysosomes [17]. Interestingly, Lys-48- and Lys-63-linked ubiquitination appears to be largely segregated to GABAB2 and GABAB1 [10, 17], respectively, which might be explained by a selective interaction of the subunits with the respective E3 ligases. However, the mechanisms regulating lysosomal degradation of GABAB receptors in response to changes in the physiological state of the neuron were unclear.
Under normal physiological conditions, cell surface GABAB receptors constitutively internalize to early endosomes and recycle to the plasma membrane or are sorted to lysosomes for degradation [12,13,14, 35, 36]. Sorting the receptors to recycling or degradation must be precisely regulated in order to provide the required number of cell surface receptors for signal transduction under a given physiological condition. However, after prolonged activation of glutamate receptors, GABAB receptors are rapidly downregulated by shifting the recycling/degradation balance towards lysosomal degradation [30,31,32]. This downregulation of GABAB receptors critically depends on CaMKII-mediated phosphorylation of GABAB1 on Ser-867 [29] as well as on Lys-63-linked ubiquitination of GABAB1 mediated by the E3 ligase MIB2 [17]. As the level of CaMKII activity is regulated by the intracellular Ca2+ concentration, it is in an ideal position to link the increased neuronal activity after prolonged activation of glutamate receptors to lysosomal degradation of GABAB receptors at least under pathological conditions. We, therefore, hypothesized that CaMKII regulates lysosomal degradation also under physiological conditions. Indeed, we found that pharmacologically blocking CaMKII significantly increased cell surface expression of GABAB receptors. This increase was accompanied by a reduced co-localization of the receptors with the late endosomal marker Rab7, indicating that inhibition of CaMKII prevented sorting of GABAB receptors to lysosomes. Our data also imply that a considerable fraction of GABAB receptors is constitutively degraded under basal conditions since blocking CaMKII activity for only 7.5 min was sufficient to induce a significant increase (~ 150%) of cell surface GABAB receptors.
CaMKII is a large protein complex constituted from 12 catalytically active subunits of different isoforms (CaMKIIα-δ) [33]. In brain, CaMKIIα and CaMKIIβ are the predominant subunits and in forebrain neurons the CaMKII holoenzyme largely consists of nine α subunits and three β subunits [37, 38]. Interestingly, we found that overexpression of CaMKIIβ, but not CaMKIIα, increased the level of GABAB receptor/CaMKII interaction in an activity-dependent manner (inhibition of CaMKII activity prevented the increase in interaction), resulting in decreased cell surface receptor expression. Consistent with this observation, overexpressing its functionally inactive mutant CaMKIIβ(DN), which was expected to inhibit CaMKIIβ activity and thus lysosomal degradation of the receptors, increased cell surface expression of GABAB receptors. As intracellular Ca2+ concentrations under basal conditions are low, this finding fits well to the observation that CaMKIIβ exhibits a ninefold higher affinity to Ca2+/calmodulin than CaMKIIα [38]. Thus, our data suggest that under normal physiological conditions basal CaMKIIβ activity determines the level of lysosomal degradation of GABAB receptors.
Phosphorylation often regulates ubiquitination of proteins, thereby promoting their degradation [39,40,41]. Several lines of evidence indicate that regulation of cell surface GABAB receptors by CaMKIIβ is conveyed by MIB2-mediated Lys-63-linked ubiquitination of GABAB1. First, overexpression in neurons of ubiquitin mutants unable to form Lys-63-linkages prevented downregulation of GABAB receptors induced by overexpression of CaMKIIβ, while a mutant that only can form Lys-63-linkages, but not any other kind of linkages, did not inhibit downregulation of the receptors. Second, blocking CaMKII activity pharmacologically or by overexpression of the functionally inactive mutant CaMKIIβ(DN) reduced Lys-63-linked ubiquitination of GABAB receptors. Third, the cell surface expression of three GABAB1a(K->R) mutants with inactivated Lys-63-linked ubiquitination sites, which are indispensable for lysosomal targeting of the receptors [17], remained unaffected by CaMKII inhibition. Fourth, overexpression of the E3 ligase MIB2, which downregulates GABAB receptors via lysosomal degradation [17], neither enhanced CaMKIIβ-mediated downregulation of cell surface receptors nor affected the expression of a GABAB1 mutant with inactivated CaMKII phosphorylation site (GABAB1(S867A)). Finally, our experiments with the CaMKII phosphorylation-deficient and phosphomimetic GABAB1(S867) mutants indicate that direct phosphorylation of GABAB1 Ser-867 promotes Lys-63-linked ubiquitination and thereby degradation of GABAB receptors. Under normal physiological as well as conditions of over-excitation via prolonged activation of glutamate receptors, the phospho-deficient mutant GABAB(S867A) displayed only marginal Lys-63-linked ubiquitination and increased cell surface expression whereas the phosphomimetic mutant GABAB1(S867D) exhibited strongly increased Lys-63-linked ubiquitination and reduced cell surface expression.
The precise mechanism by which CaMKIIβ promotes Lys63-linked ubiquitination of GABAB1 remains unresolved. Phosphorylation of GABAB1(S867) might be the signal that targets the receptors to an endosomal compartment where they are Lys-63-linked ubiquitinated by MIB2. Alternatively, phosphorylation of GABAB1(S867) might induce a conformational change in the receptor that uncovers the MIB2 interaction site or exposes the lysine residues for ubiquitination. We favor the latter hypothesis, which is supported by our finding that MIB2 displayed increased interaction with the phosphomimetic mutant GABAB1(S867D) and reduced interaction with the phospho-deficient mutant GABAB1(S867A). However, further experimentation is required to reveal the mechanism as to how CaMKIIβ promotes Lys-63-linked ubiquitination of GABAB1.
In addition to CaMKIIβ controlling cell surface GABAB receptor expression under normal physiological conditions, it rapidly downregulates the receptors upon sustained activation of glutamate receptors [29,30,31,32], a pathological condition that occurs in cerebral ischemia and leads to excitotoxic cell death [42, 43]. Downregulation of GABAB receptors is triggered by increased Ca2+ influx through NMDA receptors and voltage-gated Ca2+ channels [30], leading to enhanced CaMKII-mediated phosphorylation of GABAB1 on Ser867 [29]. This causes the re-routing of GABAB receptors from the normal constitutive recycling pathway to lysosomal degradation. Our present results show that this pathological downregulation of GABAB receptors is maintained after removal of glutamate due to the upregulation of CaMKII expression and the increased interaction of CaMKII with GABAB receptors. Therefore, interfering with this pathway to normalize GABAB receptor-mediated inhibition might be a potential approach to counteract over-excitation and to limit neuronal death.
In conclusion, our data indicate that phosphorylation of GABAB1 at Ser-867 by CaMKIIβ induces MIB2-mediated K63-linked ubiquitination of GABAB1 at multiple sites, which sorts GABAB receptors to lysosomes for degradation. This mechanism is expected to fine-tune cell surface expression of GABAB receptors under physiological conditions and to considerably affect receptor expression in diseases associated with disturbed Ca2+ homeostasis.
References
Benke D, Zemoura K, Maier PJ (2012) Modulation of cell surface GABAB receptors by desensitization, trafficking and regulated degradation. World J Biol Chem 3(4):61–72. https://doi.org/10.4331/wjbc.v3.i4.61
Pinard A, Seddik R, Bettler B (2010) GABAB receptors: physiological functions and mechanisms of diversity. Adv Pharmacol 58:231–255. https://doi.org/10.1016/s1054-3589(10)58010-4
Kohl MM, Paulsen O (2010) The roles of GABAB receptors in cortical network activity. Adv Pharmacol 58:205–229. https://doi.org/10.1016/s1054-3589(10)58009-8
Craig MT, McBain CJ (2014) The emerging role of GABAB receptors as regulators of network dynamics: fast actions from a ‘slow’ receptor? Curr Opin Neurobiol 26:15–21. https://doi.org/10.1016/j.conb.2013.10.002
Gaiarsa JL, Porcher C (2013) Emerging neurotrophic role of GABAB receptors in neuronal circuit development. Front Cell Neurosci 7:206. https://doi.org/10.3389/fncel.2013.00206
Benarroch EE (2012) GABAB receptors: structure, functions, and clinical implications. Neurology 78(8):578–584. https://doi.org/10.1212/WNL.0b013e318247cd03
Bowery NG (2006) GABAB receptor: a site of therapeutic benefit. Curr Opin Pharmacol 6(1):37–43. https://doi.org/10.1016/j.coph.2005.10.002
Cryan JF, Kaupmann K (2005) Don’t worry ‘B’ happy!: a role for GABAB receptors in anxiety and depression. Trends Pharmacol Sci 26(1):36–43
Kumar K, Sharma S, Kumar P, Deshmukh R (2013) Therapeutic potential of GABAB receptor ligands in drug addiction, anxiety, depression and other CNS disorders. Pharmacol Biochem Behav 110:174–184. https://doi.org/10.1016/j.pbb.2013.07.003
Zemoura K, Schenkel M, Acuna MA, Yevenes GE, Zeilhofer HU, Benke D (2013) Endoplasmic reticulum-associated degradation (ERAD) controls cell surface expression of γ-aminobutyric acid, type B receptors. J Biol Chem 288(48):34897–34905. https://doi.org/10.1074/jbc.M113.514745
Zemoura K, Benke D (2014) Proteasomal degradation of γ-aminobutyric acidB receptors is mediated by the interaction of the GABAB2 C terminus with the proteasomal ATPase Rtp6 and regulated by neuronal activity. J Biol Chem 289:7738–7746. https://doi.org/10.1074/jbc.M113.541987
Grampp T, Sauter K, Markovic B, Benke D (2007) γ-Aminobutyric acid type B receptors are constitutively internalized via the clathrin-dependent pathway and targeted to lysosomes for degradation. J Biol Chem 282:24157–24165. https://doi.org/10.1074/jbc.M702626200
Grampp T, Notz V, Broll I, Fischer N, Benke D (2008) Constitutive, agonist-accelerated, recycling and lysosomal degradation of GABAB receptors in cortical neurons. Mol Cell Neurosci 39(4):628–637. https://doi.org/10.1016/j.mcn.2008.09.004
Vargas KJ, Terunuma M, Tello JA, Pangalos MN, Moss SJ, Couve A (2008) The availability of surface GABAB receptors is independent of g-aminobutyric acid but controlled by glutamate in central neurons. J Biol Chem 283(36):24641–24648. https://doi.org/10.1074/jbc.M802419200
Hannan S, Wilkins ME, Dehghani-Tafti E, Thomas P, Baddeley SM, Smart TG (2011) GABAB receptor internalisation is regulated by the R2 subunit. J Biol Chem 286(27):24324–24335. https://doi.org/10.1074/jbc.M111.220814
Kantamneni S, Holman D, Wilkinson KA, Correa SA, Feligioni M, Ogden S, Fraser W, Nishimune A et al (2008) GISP binding to TSG101 increases GABAB receptor stability by down-regulating ESCRT-mediated lysosomal degradation. J Neurochem 107(1):86–95. https://doi.org/10.1111/j.1471-4159.2008.05580.x
Zemoura K, Trumpler C, Benke D (2016) Lys-63-linked ubiquitination of γ-aminobutyric acid (GABA), Type B1, at multiple sites by the E3 ligase Mind Bomb-2 targets GABAB receptors to lysosomal degradation. J Biol Chem 291:21682–21693. https://doi.org/10.1074/jbc.M116.750968
Benke D, Honer M, Michel C, Bettler B, Mohler H (1999) γ-Aminobutyric acid type B receptor splice variant proteins GBR1a and GBR1b are both associated with GBR2 in situ and display differential regional and subcellular distribution. J Biol Chem 274:27323–27330
Benke D, Michel C, Mohler H (2002) Structure of GABAB receptors in rat retina. J Recept Signal Transduc 22:253–266. https://doi.org/10.1081/RRS-120014600
Kaupmann K, Huggel K, Heid J, Flor PJ, Bischoff S, Mickel SJ, McMaster G, Angst C et al (1997) Expression cloning of GABAB receptors uncovers similarity to metabotropic glutamate receptors. Nature 386(6622):239–246. https://doi.org/10.1038/386239a0
Kaupmann K, Malitschek B, Schuler V, Heid J, Froestl W, Beck P, Mosbacher J, Bischoff S et al (1998) GABAB-receptor subtypes assemble into functional heteromeric complexes. Nature 396(6712):683–687. https://doi.org/10.1038/25360
Shen K, Teruel MN, Subramanian K, Meyer T (1998) CaMKIIbeta functions as an F-actin targeting module that localizes CaMKIIalpha/beta heterooligomers to dendritic spines. Neuron 21(3):593–606
Shen K, Meyer T (1999) Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science 284(5411):162–166
Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W et al (2005) Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci 25(8):2002–2009. https://doi.org/10.1523/JNEUROSCI.4474-04.2005
Buerli T, Pellegrino C, Baer K, Lardi-Studler B, Chudotvorova I, Fritschy JM, Medina I, Fuhrer C (2007) Efficient transfection of DNA or shRNA vectors into neurons using magnetofection. Nat Protoc 2(12):3090–3101. https://doi.org/10.1038/nprot.2007.445
Maier PJ, Zemoura K, Acuna MA, Yevenes GE, Zeilhofer HU, Benke D (2014) Ischemia-like oxygen and glucose deprivation mediates down-regulation of cell surface γ-aminobutyric acidB receptors via the endoplasmic reticulum (ER) stress-induced transcription factor CCAAT/enhancer-binding protein (C/EBP)-homologous protein (CHOP). J Biol Chem 289(18):12896–12907. https://doi.org/10.1074/jbc.M114.550517
Leuchowius KJ, Jarvius M, Wickstrom M, Rickardson L, Landegren U, Larsson R, Soderberg O, Fryknas M et al (2010) High content screening for inhibitors of protein interactions and post-translational modifications in primary cells by proximity ligation. Mol Cell Proteomics 9(1):178–183. https://doi.org/10.1074/mcp.M900331-MCP200
Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P et al (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3(12):995–1000. https://doi.org/10.1038/nmeth947
Guetg N, Aziz SA, Holbro N, Turecek R, Rose T, Seddik R, Gassmann M, Moes S et al (2010) NMDA receptor-dependent GABAB receptor internalization via CaMKII phosphorylation of serine 867 in GABAB1. Proc Natl Acad Sci U S A 107(31):13924–13929. https://doi.org/10.1073/pnas.1000909107
Maier PJ, Marin I, Grampp T, Sommer A, Benke D (2010) Sustained glutamate receptor activation down-regulates GABAB receptors by shifting the balance from recycling to lysosomal degradation. J Biol Chem 285(46):35606–35614. https://doi.org/10.1074/jbc.M110.142406
Terunuma M, Vargas KJ, Wilkins ME, Ramirez OA, Jaureguiberry-Bravo M, Pangalos MN, Smart TG, Moss SJ et al (2010) Prolonged activation of NMDA receptors promotes dephosphorylation and alters postendocytic sorting of GABAB receptors. Proc Natl Acad Sci U S A 107(31):13918–13923. https://doi.org/10.1073/pnas.1000853107
Kantamneni S, Gonzalez-Gonzalez IM, Luo J, Cimarosti H, Jacobs SC, Jaafari N, Henley JM (2014) Differential regulation of GABAB receptor trafficking by different modes of N-methyl-D-aspartate (NMDA) receptor signaling. J Biol Chem 289(10):6681–6694. https://doi.org/10.1074/jbc.M113.487348
Hell JW (2014) CaMKII: claiming center stage in postsynaptic function and organization. Neuron 81(2):249–265. https://doi.org/10.1016/j.neuron.2013.12.024
Ng EL, Gan BQ, Ng F, Tang BL (2012) Rab GTPases regulating receptor trafficking at the late endosome-lysosome membranes. Cell Biochem Funct 30(6):515–523. https://doi.org/10.1002/cbf.2827
Pooler AM, Gray AG, McIlhinney RA (2009) Identification of a novel region of the GABAB2 C-terminus that regulates surface expression and neuronal targeting of the GABAB receptor. Eur J Neurosci 29(5):869–878. https://doi.org/10.1111/j.1460-9568.2009.06636.x
Wilkins ME, Li X, Smart TG (2008) Tracking cell surface GABAB receptors using an α-bungarotoxin tag. J Biol Chem 283(50):34745–34752. https://doi.org/10.1074/jbc.M803197200
Miller SG, Kennedy MB (1985) Distinct forebrain and cerebellar isozymes of type II Ca2+/calmodulin-dependent protein kinase associate differently with the postsynaptic density fraction. J Biol Chem 260(15):9039–9046
Brocke L, Chiang LW, Wagner PD, Schulman H (1999) Functional implications of the subunit composition of neuronal CaM kinase II. J Biol Chem 274(32):22713–22722
Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C (2002) Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J 21(15):4037–4048
Schroeder B, Srivatsan S, Shaw A, Billadeau D, McNiven MA (2012) CIN85 phosphorylation is essential for EGFR ubiquitination and sorting into multivesicular bodies. Mol Biol Cell 23(18):3602–3611. https://doi.org/10.1091/mbc.E11-08-0666
Su H, Chen M, Sands JM, Chen G (2013) Activation of the cAMP/PKA pathway induces UT-A1 urea transporter monoubiquitination and targets it for lysosomal degradation. Am J Physiol Renal Physiol 305(12):F1775–F1782. https://doi.org/10.1152/ajprenal.00393.2013
Kostandy BB (2012) The role of glutamate in neuronal ischemic injury: the role of spark in fire. Neurol Sci 33(2):223–237. https://doi.org/10.1007/s10072-011-0828-5
Lipton P (1999) Ischemic cell death in brain neurons. Physiol Rev 79(4):1431–1568. https://doi.org/10.1152/physrev.1999.79.4.1431
Acknowledgements
We are grateful to Giovanna Bosshard for dissecting the E18 rat cortex.
Funding
This study was supported by the Swiss National Science Foundation (grant 31003A_138382 to D.B.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zemoura, K., Balakrishnan, K., Grampp, T. et al. Ca2+/Calmodulin-Dependent Protein Kinase II (CaMKII) β-Dependent Phosphorylation of GABAB1 Triggers Lysosomal Degradation of GABAB Receptors via Mind Bomb-2 (MIB2)-Mediated Lys-63-Linked Ubiquitination. Mol Neurobiol 56, 1293–1309 (2019). https://doi.org/10.1007/s12035-018-1142-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-018-1142-5