Abstract
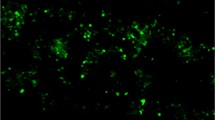
The synthesis of brain metabolic DNA (BMD) is modulated by learning and circadian oscillations and is not involved in cell division or DNA repair. Data from rats have highlighted its prevalent association with the mitochondrial fraction and its lack of identity with mtDNA. These features suggested that BMD could be localized in synaptosomes that are the major contaminants of brain mitochondrial fractions. The hypothesis has been examined by immunochemical analyses of the large synaptosomes of squid optic lobes that are readily prepared and identified. Optic lobe slices were incubated with 5-bromo-2-deoxyuridine (BrdU) and the isolated synaptosomal fraction was exposed to the green fluorescent anti-BrdU antibody. This procedure revealed that newly synthesized BrdU-labeled BMD is present in a significant percent of the large synaptosomes derived from the nerve terminals of retinal photoreceptor neurons and in synaptosomal bodies of smaller size. Synaptosomal BMD synthesis was strongly inhibited by actinomycin D. In addition, treatment of the synaptosomal fraction with Hoechst 33258, a blue fluorescent dye specific for dsDNA, indicated that native DNA was present in all synaptosomes. The possible role of synaptic BMD is briefly discussed.



Similar content being viewed by others
References
Pelc SR (1964) Labelling of DNA and cell division in so called non-dividing tissues. J Cell Biol 22:21–28
Pelc SR (1968) Turnover of DNA and function. Nature 219:162–l63
Pelc SR (1968) Biological implications of DNA-turnover in higher organisms. Acta Histochem Suppl 8:441–452
Pelc SR (1972) Metabolic DNA in ciliated protozoa, salivary gland chromosomes, and mammalian cells. Int Rev Cytol 32:327–355
Pelc SR, Viola-Magni MP (1969) Decrease of labeled DNA in cells of the adrenal medulla after intermittent exposure to cold. J Cell Biol 42:460–468
Roels H (1966) “Metabolic” DNA: a cytochemical study. Int Rev Cytol 19:1–34
Stroun M, Charles P, Anker P, Pelc SR (1967) Metabolic DNA in heart and skeletal muscle and in the intestine of mice. Nature 216:716–717
Gahan PB, Anker P, Stroun M (2008) Metabolic DNA as the origin of spontaneously released DNA? Ann N Y Acad Sci 1137:7–17
Perrone Capano C, D'Onofrio G, Giuditta A (1982) DNA turnover in rat cerebral cortex. J Neurochem 38:52–56
Reinis S (1972) Autoradiographic study of 3H-thymidine incorporation into brain DNA during learning. Physiol Chem Phys 4:391–397
Reinis S, Lamble RW (1972) Labeling of brain DNA by 3H-thymidine during learning. Physiol Chem Phys 4:335–338
Ashapkin VV, Romanov GA, Tushmalova NA, Vanyushin BF (1983) Selective DNA synthesis in the rat brain induced by learning. Biokhimija 48:355–362
Scaroni R, Ambrosini MV, Principato GB, Federici F, Ambrosi G, Giuditta A (1983) Synthesis of brain DNA during acquisition of an active avoidance task. Physiol Behav 30:577–582
Giuditta A, Perrone Capano C, D'Onofrio G, Toniatti C, Menna T, Hydèn H (1986) Synthesis of rat brain DNA during acquisition of an appetitive task. Pharmacol Biochem Behav 25:651–658
Giuditta A, Ambrosini MV, Scaroni R, Chiurulla C, Sadile A (1985) Effect of sleep on cerebral DNA synthesized during shuttle-box avoidance training. Physiol Behav 34:769–778
Langella M, Colarieti L, Ambrosini MV, Giuditta A (1992) The sequential hypothesis of sleep function. IV. A correlative analysis of sleep variables in learning and nonlearning rats. Physiol Behav 51:227–238
Grassi Zucconi G, Menichini E, Castigli E, Belia S, Giuditta A (1988a) Circadian oscillations of DNA synthesis in rat brain. Brain Res 447:253–261
Grassi Zucconi G, Carandente ME, Belia S, Giuditta A (1988b) Circadian rhythms of DNA content in brain and kidney: effects of environmental stimulation. Chronobiol 15:195–204
Grassi Zucconi G, Crognale MC, Bassetti MA, Giuditta (1990) Environmental stimuli modulate the circadian rhythm of (3H-methyl)thymidine incorporation into brain DNA of male rats. Behav Brain Res 41:103–110
Giuditta A, Grassi-Zucconi G, Sadile A (2017) Brain metabolic DNA in memory processing and genome turnover. Rev Neurosci 28:21–30
Rutigliano B, Giuditta A (2015) The unexpected recovery of misplaced data on brain metabolic DNA. Rend Acc Sci Fis Mat LXXXII:99–106
Giuditta A, Rutigliano B (2017) Brain metabolic DNA in rat cytoplasm. Mol Neurobiol. https://doi.org/10.1007/s12035-018-0932-0
Satake M, Abe S (1966) Preparation and characterization of nerve cell perikaryon from rat cerebral cortex. J Biochem 59:72–75
Crispino M, Castigli E, Perrone Capano C, Martin, Menichini E, Kaplan BB, Giuditta A (1993) Protein synthesis in a synaptosomal fraction from squid brain. Mol R Cell Neurosci 4:366–374
Eyman M, Cefaliello C, Ferrara E, De Stefano R, Scotto Lavina Z, Crispino M, Squillace A, van Minnen J et al (2007) Local synthesis of axonal and presynaptic RNA in squid model systems. Eur J Neurosci 25:341–350
Cefaliello C, Prisco M, Crispino M, Giuditta A (2015) Newly synthesized DNA in squid nerve terminals. Rend Acc Sc Fis Mat Napoli LXXXII:61–64
Cohen RS, Pant HC, Gainer H (1990) Squid optic lobe synaptosomes: Structure and function of isolated synapses. In: Gilbert DL, Adelman Jr WJ, Arnold JM (eds) Squid as experimental animals. Plenum Press, New York, pp. 213–233
Crispino M, Kaplan BB, Martin R, Alvarez J, Chun JT, Giuditta (1997) Active polysomes are present in the large presynaptic endings of the synaptosomal fraction from squid brain. J Neurosci 17:7694–7702
Giuditta A, Chun JT, Eyman M, Cefaliello C, Crispino M (2008) Local gene expression in axons and nerve endings: the glia-neuron unit. Physiol Rev 88:515–555
Crispino M, Chun JT, Cefaliello C, Perrone Capano C, Giuditta A (2014) Local gene expression in nerve endings. Dev Neurobiol 74:279–291
Rabinovitch M, Plaut W (1962) Cytoplasmic DNA synthesis in Amoeba proteus. I. On the particulate nature of the DNA-containing elements. J Cell Biol 15:525–534
Hotta Y, Bassel A, Stern H (1965) Nuclear DNA and cytoplasmic DNA from tissues of higher plants. J Cell Biol 27:451–457
Bond HE, Cooper JA 2nd, Courington DP, Wood JS (1969) Microsome-associated DNA. Science 165:705–706
Baril EF, Jenkins MD, Brown OE, Laszlo J (1970) DNA polymerase activities associated with smooth membranes and ribosomes from rat liver and hepatoma cytoplasm. Science 169:87–89
Schneider WC (1977) Cytoplasmic DNA: differentiation by reassociation kinetics. Biochem Biophys Res Commun 74:1607–1612
Williams RP, Wilhelm GB, Adrian EK Jr, Cameron IL (1982) Evidence for turnover of DNA in the nucleus and in the cytoplasm of adult post-mitotic neurons in vivo. Cytobios 35:187–194
Koch J, Vogt G, Kissel W (1983) Cytoplasmic DNA is structurally different from nuclear DNA. Naturwissenschaften 70:252–254
Cimarra P, Giuditta A (1979) Identification of a group of octopus brain proteins as histones. J Neurochem 32(12):1279–1286
Salganik RI, Parvez H, Tomson VP, Shumskaya IA (1983) Probable role of reverse transcription in learning: correlation between hippocampal RNA-dependent DNA synthesis learning ability in rats. Neurosci Lett 36:317–322
Montagnier L, Del Giudice E, Aïssa J et al (2015) Transduction of DNA information through water and electromagnetic waves. Electromagn Biol Med 34:106–112
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
All authors declare that they have no conflict of interests.
Additional information
Antonio Giuditta is emeritus professor of Physiology
Rights and permissions
About this article
Cite this article
Cefaliello, C., Prisco, M., Crispino, M. et al. DNA in Squid Synaptosomes. Mol Neurobiol 56, 56–60 (2019). https://doi.org/10.1007/s12035-018-1071-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-018-1071-3