Abstract
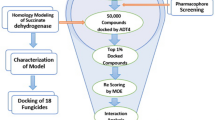
Fusarium oxysporum f. sp. cubense is one of the most severe and threatening pathogens of bananas, causing “Panama wilt” worldwide. Confrontation assay of Foc antagonistic bacterial endophyte, Bacillus velezensis YEBBR6, with the Foc and GC–MS profiling of excised agar from the zone of inhibition, led to the unveiling of secondary metabolites produced by the endophyte. To refine the probable antifungal compounds among the numerous biomolecules formed during their di-trophic interaction with the pathogen, fungal protein targets were modeled, and docking studies (AutoDock Vina module of the PyRx 0.8 server) were done with all the compounds. Triamcinolone acetonide exhibited the most excellent affinity for the protein targets among the compounds studied. It had a maximum binding affinity of 11.2 kcal/mol for XRN2 (5ʹ → 3ʹ). Further, the protein–ligand complex formation kinetics was done through Molecular Dynamic Simulation studies. Graphs for the RMSD, RMSF, Rg, potential energy, and SASA were generated, and the values during the simulation period suggested the stability of the biomolecule as a complex with the protein. This indicated Triamcinolone acetonide’s potential ability to act as a functional disrupter of the target protein and likely an antifungal molecule. Further, the biomolecule was tested for its activity against Foc by screening in the wet lab through the poisoned plate technique, and it was found to be fully inhibitory to the growth of the pathogen at 1000 ppm.













Similar content being viewed by others
Data availability
The present study’s data are included in the manuscript and its supplementary file.
References
FAO. (2021). Banana Market Review Preliminary Results 2020, Vol. 2022. Rome
FAO. (2022). Banana market review – Preliminary results 2021, Vol. 2022. Rome, Cjam
Srabani, G., Shri Anil Kumar, S., Ramesh Kumar, Y., Shri Ashutosh, S., Shri Manish, Y., Neha, A., Shri Subhash, D., & Shri Gautam, P. (2021). Agricultural Statistics at a Glance 2020, Vol. 2022, Government of India, Ministry of Agriculture & Farmers Welfare, Department of Agriculture, Cooperation & Farmers Welfare, Directorate of Economics and Statistics.
Stover, R.H. (1962). Fusarial wilt (Panama disease) of bananas and other Musa species. Commonwealth Mycological Institute, Kew, England.
Ploetz, R. C. (2006). Fusarium Wilt of Banana Is Caused by Several Pathogens Referred to as Fusarium oxysporum f. sp. cubense. Phytopathology, 96(6), 653–656.
Ploetz, R. C. (2006). Panama Disease: An Old Nemesis Rears its Ugly: Head Part 2. The Cavendish Era and Beyond. Plant Health Progress, 7(1), 36.
Maymon, M., Sela, N., Shpatz, U., Galpaz, N., & Freeman, S. (2020). The origin and current situation of Fusarium oxysporum f. sp. cubense tropical race 4 in Israel and the Middle East. Scientific Reports, 10(1), 1590.
Ploetz, R., & Pegg, K. (1997). Fusarium wilt of banana and Wallace’s line: Was the disease originally restricted to his Indo-Malayan region? Australasian Plant Pathology, 26(4), 239–249.
Ploetz, R. C. (2015). Management of Fusarium wilt of banana: A review with special reference to tropical race 4. Crop Protection, 73, 7–15.
Dita, M., Barquero, M., Heck, D., Mizubuti, E. S. G., & Staver, C. P. (2018). Fusarium Wilt of Banana: Current knowledge on epidemiology and research needs toward sustainable disease management. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2018.01468
Pegg, K. G., Coates, L. M., O’Neill, W. T., & Turner, D. W. (2019). The Epidemiology of Fusarium Wilt of Banana. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2019.01395
Guo, G., Wang, B., Ma, W., Li, X., Zhu, C., Ming, J., & Zeng, H. (2013). Biocontrol of Fusarium wilt of banana: Key influence factors and strategies. African Journal of Microbiology Research, 7(41), 4835–4843.
Wei, Y., Liu, W., Hu, W., Liu, G., Wu, C., Liu, W., Zeng, H., He, C., & Shi, H. (2017). Genome-wide analysis of autophagy-related genes in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Reports, 36(8), 1237–1250.
Mahdi, M. A., Yousefi, S. R., Jasim, L. S., & Salavati-Niasari, M. (2022). Green synthesis of DyBa2Fe3O7. 988/DyFeO3 nanocomposites using almond extract with dual eco-friendly applications: Photocatalytic and antibacterial activities. International Journal of Hydrogen Energy, 47(31), 14319–14330.
Yousefi, S. R., Alshamsi, H. A., Amiri, O., & Salavati-Niasari, M. (2021). Synthesis, characterization and application of Co/Co3O4 nanocomposites as an effective photocatalyst for discoloration of organic dye contaminants in wastewater and antibacterial properties. Journal of Molecular Liquids, 337, 116405.
Amer, G. A., & Utkhede, R. S. (2000). Development of formulations of biological agents for management of lettuce and cucumber root rot. Canadian Journal of Microbiology, 46(9), 809–816.
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., & Dowling, D. N. (2008). Bacterial endophytes: Recent developments and applications. FEMS Microbiology Letters, 278(1), 1–9.
Bhattacharyya, P. N., & Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World Journal of Microbiology & Biotechnology, 28(4), 1327–1350.
Ravi, S., Sevugapperumal, N., Nallusamy, S., Shanmugam, H., Mathiyazhagan, K., Rangasamy, A., Akkanna Subbiah, K., & Varagur Ganesan, M. (2022). Differential bacterial endophytome in Foc-resistant banana cultivar displays enhanced antagonistic activity against Fusarium oxysporum f.sp. cubense (Foc). Environmental Microbiology., 24(6), 2701–2715.
Ali, S. A. M., Sayyed, R. Z., Mir, M. I., Hameeda, B., Khan, Y., Alkhanani, M. F., Haque, S., & Tawaha, A. R. M. A. (2022). Induction of Systemic Resistance and Antibiofilm activity of Surfactin from Bacillus velezensis MS20 and evaluation of its Induced. Frontiers in Microbiology., 13, 879739. https://doi.org/10.3389/fmicb.2022.879739
Debois, D., Hamze, K., Guérineau, V., Le Caër, J. P., Holland, I. B., Lopes, P., Ouazzani, J., Séror, S. J., Brunelle, A., & Laprévote, O. (2008). In situ localisation and quantification of surfactins in a Bacillus subtilis swarming community by imaging mass spectrometry. Proteomics, 8(18), 3682–3691.
Jadhav, H. P., Sayyed, R. Z., Shaikh, S. S., Bhamare, H. M., Sunita, K., & Enshasy, H. E. (2020). Statistically Designed Bioprocess for Enhanced Production of Alkaline Protease in Bacillus cereus HP_RZ17. Journal of Scientific & Industrial Research, 79, 491–498.
Raaijmakers, J. M., De Bruijn, I., Nybroe, O, & Ongena, M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiology Reviews 34(6), 1037-1062
Sagar, A., Yadav, S. S., Sayyed, R. Z., Sharma, S., & Ramteke, P. W. (2022). Bacillus subtilis: A Multifarious Plant Growth Promoter, Biocontrol Agent, and Bioalleviator of Abiotic Stress, Bacilli in Agrobiotechnology. In M. T. Islam, M. Rahman, & P. Pandey (Eds.), Bacilli in Agrobiotechnology Bacilli in Climate Resilient Agriculture and Bioprospecting (pp. 561–580). Springer.
Mora, I., Cabrefiga, J., & Montesinos, E. (2011). Antimicrobial peptide genes in Bacillus strains from plant environments. International Microbiology, 14(4), 213–223.
Nakkeeran, S., Rajamanickam, S., Saravanan, R., Vanthana, M., & Soorianathasundaram, K. (2021). Bacterial endophytome-mediated resistance in banana for the management of Fusarium wilt. 3 Biotech, 11(6), 1–13.
Guedes, I. A., de Magalhães, C. S., & Dardenne, L. E. (2014). Receptor–ligand molecular docking. Biophysical reviews, 6(1), 75–87.
Dar, A. M., & Mir, S. (2017). Molecular docking: Approaches, types, applications and basic challenges. Journal of Analytical & Bioanalytical, 8(2), 1–3.
Huang, Y.-mM., Kang, M., & Chang, C.-eA. (2014). Switches of hydrogen bonds during ligand–protein association processes determine binding kinetics. Journal of Molecular Recognition, 27(9), 537–548.
Du, X., Li, Y., Xia, Y.-L., Ai, S.-M., Liang, J., Sang, P., Ji, X.-L., & Liu, S.-Q. (2016). Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. International Journal of Molecular Sciences, 17(2), 144.
Dutta Dubey, K., Kumar Tiwari, R., & Prasad Ojha, R. (2013). Recent advances in protein− ligand interactions: Molecular dynamics simulations and binding free energy. Current Computer Aided-Drug Design, 9(4), 518–531.
Soundararajan, P., Sakkiah, S., Sivanesan, I., Lee, K.-W., & Jeong, B.-R. (2011). Macromolecular docking simulation to identify binding site of FGB1 for antifungal compounds. Bulletin of the Korean Chemical Society, 32(10), 3675–3681.
Gurdaswani, V., Ghag, S. B., & Ganapathi, T. R. (2020). FocSge1 in Fusarium oxysporum f. sp. cubense race 1 is essential for full virulence. BMC Microbiology, 20(1), 255.
Martínez-Rocha, A. L., Roncero, M. I. G., López-Ramirez, A., Mariné, M., Guarro, J., Martínez-Cadena, G., & Di Pietro, A. (2008). Rho1 has distinct functions in morphogenesis, cell wall biosynthesis and virulence of Fusarium oxysporum. Cellular Microbiology, 10(6), 1339–1351.
Maldonado Bonilla, L., & Calderón-Oropeza, M. (2018). The 5→ 3 Exoribonuclease 2 as a Potential Target for Developing Fungicides to Control the Panama Disease. Journal of Plant Pathology & Microbiology., 9(453), 2.
Deng, G.-M., Yang, Q.-S., He, W.-D., Li, C.-Y., Yang, J., Zuo, C.-W., Gao, J., Sheng, O., Lu, S.-Y., Zhang, S., & Yi, G.-J. (2015). Proteomic analysis of conidia germination in Fusarium oxysporum f. sp. cubense tropical race 4 reveals new targets in ergosterol biosynthesis pathway for controlling Fusarium wilt of banana. Applied Microbiology and Biotechnology., 99(17), 7189–7207.
Ghag, S. B., Shekhawat, U. K. S., & Ganapathi, T. R. (2014). Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnology Journal, 12(5), 541–553.
Ding, Z., Xu, T., Zhu, W., Li, L., & Fu, Q. (2020). A MADS-box transcription factor FoRlm1 regulates aerial hyphal growth, oxidative stress, cell wall biosynthesis and virulence in Fusarium oxysporum f. sp. cubense. Fungal Biology, 124(3), 183–193.
Ding, Z., Li, M., Sun, F., Xi, P., Sun, L., Zhang, L., & Jiang, Z. (2015). Mitogen-activated protein kinases are associated with the regulation of Physiological Traits and Virulence in Fusarium oxysporum f. sp. cubense. PLoS one, 10(4), e0122634.
Carvalhais, L. C., Henderson, J., Rincon-Florez, V. A., O’Dwyer, C., Czislowski, E., Aitken, E. A. B., & Drenth, A. (2019). Molecular Diagnostics of Banana Fusarium Wilt Targeting Secreted-in-Xylem Genes. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2019.00547
Rehman, I., Kerndt, C. C., & Botelho, S. (2022). Biochemistry Tertiary Protein Structure. StatPearls Publishing.
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., Martin, M. J., Natale, D. A., O’Donovan, C., Redaschi, N., & Yeh, L. S. L. (2004). UniProt: the Universal Protein knowledgebase. Nucleic Acids Research, 32(suppl_1), D115–D119.
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., & Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Research, 36(suppl_2), W5–W9.
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F. T., de Beer, T. A. P., Rempfer, C., Bordoli, L., Lepore, R., & Schwede, T. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), W296–W303.
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., & Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 10(6), 845–858.
Kim, D. E., Chivian, D., & Baker, D. (2004). Protein structure prediction and analysis using the Robetta server. Nucleic Acids Research, 32(2), W526–W531.
Laskowski, R. A., MacArthur, M. W., Moss, D. S., & Thornton, J. M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26(2), 283–291.
Binkowski, T. A., Naghibzadeh, S., & Liang, J. (2003). CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Research, 31(13), 3352–3355.
Dallakyan, S., & Olson, A. J. (2015). Small-Molecule Library Screening by Docking with PyRx. In J. E. Hempel, C. H. Williams, & C. C. Hong (Eds.), Chemical Biology: Methods and Protocols (pp. 243–250). Springer.
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). saiGROMACS: High-performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25.
Meng, X.-Y., Zhang, H.-X., Mezei, M., & Cui, M. (2011). Molecular docking: A powerful approach for structure-based drug discovery. Current Computer Aided-Drug Design, 7(2), 146–157.
Backman, T. W. H., Cao, Y., & Girke, T. (2011). ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Research, 39(2), W486–W491.
Cao, Y., Jiang, T., & Girke, T. (2008). A maximum common substructure-based algorithm for searching and predicting drug-like compounds. Bioinformatics, 24(13), i366–i374.
Kufareva, I., & Abagyan, R. (2012). Methods of protein structure comparison. Methods in Molecular Biology, 857, 231–257.
Benson, N. C., & Daggett, V. (2012). A comparison of multiscale methods for the analysis of molecular dynamics simulations. The Journal of Physical Chemistry B, 116, 8722–8731.
Schiebel, J., Gaspari, R., Wulsdorf, T., et al. (2018). Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes. Nature Communications, 9, 3559.
Marsh, J. A., & Teichmann, S. A. (2011). Relative solvent accessible surface area predicts protein conformational changes upon binding. Structure, 19(6), 859–867.
Zaki, A. A., Ashour, A., Elhady, S. S., Darwish, K. M., & Al-Karmalawy, A. A. (2022). Calendulaglycoside A showing potential activity against SARS-CoV-2 main protease: Molecular docking, molecular dynamics, and SAR studies. Journal of Traditional & Complementary Medicine, 2(1), 16–34.
Bacon, C. W., & Hinton, D. M. (2006). Bacterial endophytes: The endophytic niche, its occupants, and its utility. In S. S. Gnanamanickam (Ed.), Plant-Associated Bacteria (pp. 155–194). Springer.
Sessitsch, A., Howieson, J. G., Perret, X., Antoun, H., & Martínez-Romero, E. (2002). Advances in Rhizobium Research. Critical Reviews in Plant Sciences, 21(4), 323–378.
Sturz, A. V., Christie, B. R., & Nowak, J. (2000). Bacterial endophytes: Potential role in developing sustainable systems of crop production. Critical Reviews in Plant Sciences, 19(1), 1–30.
Kilani-Feki, O., & Jaoua, S. (2011). Biological control of Botrytis cinerea using the antagonistic and endophytic Burkholderia cepacia Cs5 for vine plantlet protection. Canadian Journal of Microbiology, 57(11), 896–901.
Salsbury, F. R. (2010). Molecular dynamics simulations of protein dynamics and their relevance to drug discovery. Current Opinion in Pharmacology, 10(6), 738–744.
Ravindranath, P. A., Forli, S., Goodsell, D. S., Olson, A.J., & Sanner, M. F. (2015). AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Computational Biology 2, 11(12), e1004586
Miki, T. S., & Großhans, H. (2013). The multifunctional RNase XRN2. Biochemical Society Transactions, 41(4), 825–830.
Petfalski, E., Dandekar, T., Henry, Y., & Tollervey, D. (1998). Processing of the Precursors to Small Nucleolar RNAs and rRNAs Requires Common Components. Molecular and Cellular Biology, 18(3), 1181–1189.
Mansouri, N., & Benslama, O. (2022). In vitro and in silico investigation of the antifungal activity of endophytic fungi against phytopathogenic fungi of tomato. Notulae Scientia Biologicae, 14(1), 11050–11050.
Mansouri, N., Benslama, O., & Arhab, R. (2021). "Homology modeling, docking and molecular dynamics studies of some secondary metabolites of actinomycetes as biocontrol agents against the 3HNR enzyme of the phytopathogenic fungus Alternaria alternata. Journal of Biomolecular Structure and Dynamics, 41(3), 871–883.
Sonkar, P., & Odumosu, B. T. (2022). Genome mining and In-silico determination of secondary metabolites of bacteria against Fusarium oxysporum f. sp. Lycopersici. Journal of Phytopathology, 170(2), 100–106.
Islam, M. N., Ali, M. S., Choi, S.-J., Hyun, J.-W., & Baek, K.-H. (2019). Biocontrol of citrus canker disease caused by Xanthomonas citri subsp. citri using an endophytic Bacillus thuringiensis. The plant pathology journal, 35(5), 486.
Angarita-Rodríguez, A., Quiroga, D., & Coy-Barrera, E. (2019). Indole-Containing Phytoalexin-Based Bioisosteres as Antifungals. In Vitro and In Silico Evaluation against Fusarium oxysporum. Molecules, 25(1), 45.
Salehi, F., Emami, L., Rezaei, Z., Khabnadideh, S., Tajik, B., & Sabet, R. (2022). Fluconazole-Like compounds as potential antifungal agents: QSAR, molecular docking, and molecular dynamics simulation. Journal of Chemistry, 2022, 1–16.
Maheen, S., Younis, H., Khan, H. U., Ali, S., Rehman, A. U., Ilyas, S., Zafar, M. N., Shafqat, S. R., Kalam, A., & Al-Ghamdi, A. A. (2022). Enhanced Antifungal and Wound Healing Efficacy of Statistically Optimized, Physicochemically Evaluated Econazole-Triamcinolone Loaded Silica Nanoparticles. Frontiers in Chemistry. https://doi.org/10.3389/fchem.2022.836678
Saravanan, R., Nakkeeran, S., Sarayna, S., Senthilraja, C., Renukadevi, P., Krishnamoorthy, A. S., Enshasy, H. E., Eldawi, H. A., Malathi, V. G., Salmen, S. H., Ansari, M. J., Khan, N., & Sayyed, R. Z. (2021). Mining the Genome of Bacillus velezensis VB7 (CP047587) for MAMP genes and non-ribosomal peptide synthetase gene clusters conferring antiviral and antifungal activity. Microorganisms, 9, 2511.
Kadiri, M., Sevugapperumal, N., Nallusamy, S., Ragunathan, J., Ganesan, M. G., Alfarraj, S., Ansari, M. J., Sayyed, R. Z., Lim, H. R., & Show, P. L. (2023). Pan-genome analysis and molecular docking unveil the biocontrol potential of Bacillus velezensis VB7 against Phytophthora infestans. Microbiological Research, 268, 127277. https://doi.org/10.1016/j.micres.2022.127277
Acknowledgements
This work was supported by the DBT–BTIS facility available at the Department of Plant Molecular Biology and Bioinformatics, Centre for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, India. The authors would like to acknowledge the support provided by Researchers Supporting Project Number RSP2023R358, King Saud University, Riyadh, Saudi Arabia. The authors acknowledge the Department of Plant Biotechnology, Department of Plant Pathology, and Department of Nanotechnology, Tamil Nadu Agricultural University, Coimbatore, India, for providing facilities.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nayana, R.U.K., Nakkeeran, S., Saranya, N. et al. Triamcinolone Acetonide Produced by Bacillus velezensis YEBBR6 Exerts Antagonistic Activity Against Fusarium oxysporum f. sp. Cubense: A Computational Analysis. Mol Biotechnol (2023). https://doi.org/10.1007/s12033-023-00797-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-023-00797-w