Abstract
An ongoing pandemic of coronavirus disease 2019 (COVID-19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). So far, there have been various approaches for SARS-CoV-2 detection, each having its pros and cons. The current gold-standard method for SARS-CoV-2 detection, which offers acceptable specificity and sensitivity, is the quantitative reverse transcription-PCR (qRT-PCR). However, this method requires considerable cost and time to transport samples to specialized laboratories and extract, amplify, and detect the viral genome. On the other hand, antigen and antibody testing approaches that bring rapidity and affordability into play have lower sensitivity and specificity during the early stages of COVID-19. Moreover, the immune response is variable depending on the individual. Methods based on clustered regularly interspaced short palindromic repeats (CRISPR) can be used as an alternative approach to controlling the spread of disease by a high-sensitive, specific, and low-cost molecular diagnostic system. CRISPR-based detection systems (CRISPR-Dx) target the desired sequences by specific CRISPR-RNA (crRNA)-pairing on a pre-amplified sample and a subsequent collateral cleavage. In the present article, we have reviewed different CRISPR-Dx methods and presented their benefits and drawbacks for point-of-care testing (POCT) of suspected SARS-CoV-2 infections at home or in small clinics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
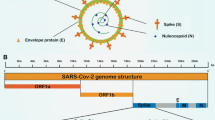
Emerging infectious diseases are challenging the healthcare systems and millions of people worldwide. In December 2019, an outbreak of the novel severe acute respiratory syndrome-related coronavirus SARS-CoV-2 (COVID-19) developed in Wuhan, China [1]. Since then, the COVID-19 outbreak has rapidly spread worldwide and is now considered a global pandemic [2]. SARS-CoV-2 has a positive-stranded RNA with a genome size ranging from ~ 27 to ~ 32 kb [3]. This virus infects various of avian and mammalian species and rapidly modifies the genome by recombination to generate more virulent and recalcitrant strains than the original strains [4, 5]. This fact is demonstrated by the recent outbreak of the novel SARS-CoV-2. The COVID-19 pandemic has posed additional socioeconomic challenges to health systems [6]. Countries have implemented strict measures to control the viral spread through social distancing. In the absence of a specific treatment for this disease, effective screening methods play a significant role in controlling the pandemic [7].
Large-scale screening is crucial for the early detection of SARS-CoV-2 infection. On-time assessment of the outbreaks is complex because of the facile transmissibility and delay in clinical manifestation of SARS-CoV-2 [8]. In the lack of robust detection tests, rapid circulation of the virus occurs within a population [9]. Nucleic acid amplification-based molecular diagnostics (MDx) are relatively helpful, accurate, fast, and affordable compared to serological approaches to screen for the current presence of the virus in the early stages of infection [10]. Currently, the gold-standard test for molecular diagnosis of COVID-19 is quantitative reverse transcription-PCR (qRT-PCR) [11]. This technique has been suggested by both the Centers for Disease Control and Prevention (CDC) [12] and the World Health Organization (WHO) [13]. However, qRT-PCR has brought urgent challenges for large-scale point-of-care diagnostics since it requires trained operators, transport of samples to central laboratories, and sophisticated laboratory infrastructure [14]. When thousands of samples must be quickly analyzed in the SARS-CoV-2 pandemic, it is needed to accurately read the results [15]. There are uncertain positive or negative rRT-PCR readouts in correlation with the frequently detected “gray zones” characterized by high Ct values [16,17,18,19]. The major intrinsic causative factors for high Ct-Values and inaccurate rRT-PCR readouts are user errors such as inaccurate sampling, unqualified reagents, uncalibrated high-tech equipment, inefficient RT reactions, and PCR amplification of patient specimens with very low viral titers. These issues result in false-negative or false-positive diagnoses of the low viral load samples from mild, asymptomatic, or recovering patients, which may raise concerns about managing the disease [20,21,22].
In emergencies, point-of-care (POC) diagnostic approaches are suggested to be adapted to the ASSURED features (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end-users) [23]. In low-resource settings, a technique named loop-mediated isothermal amplification (LAMP) can display an effective alternative to PCR-based methods [24]. LAMP contains a polymerase amplification at a single temperature, and reverse transcription LAMP (RT-LAMP) coupled with an early reverse transcription of a specific RNA sequence to DNA. Although RT-LAMP can detect RNA SARS-CoV-2 by a single reaction at a single temperature, the results show low specificity and pose difficulties in adapting it to effective POC diagnostics [25].
Many health care systems have transitioned to the era of precise molecular diagnosis using clustered regularly interspaced short palindromic repeats (CRISPR) and associated CRISPR protein (Cas) (CRISPR-Cas) [26,27,28,29,30]. CRISPR-based diagnostics (CRISPR-Dx) have developed at an extraordinary pace for rapid nucleic acid detection of virtually any DNA or RNA sequence [31]. Recently, Cas endonucleases have been used for the rapid, specific, and sensitive detection of nucleic acids [32,33,34]. These approaches rely on CRISPR-associated proteins, including Cas13 [35] or Cas12 [36], as non-specific endonucleases. If the programmed crRNAs recognize specific targets, Cas13 or Cas12 cleave the reporter molecules and produce a specific and sensitive indicator for the presence or quantity of nucleic acid [37]. Cas nucleases identify targets by making a ribonucleoprotein (RNP) with an RNA sequence called guide RNA (gRNA) [29, 38]. The gRNA includes a programmable molecule called CRISPR-RNA (crRNA) for a specific target. Moreover, it contains a non-coding RNA fragment that facilitates interactions with effector proteins through hydrogen bonding and aromatic stacking [39]. When gRNA and the effector proteins interact, an RNP surveillance complex creates to scan nucleic acids and cleave complementary crRNA sequences [40].
Thus, hybridizing gRNA can be reprogrammed and designed to target any desired downstream gene. CRISPR/Cas systems categorize into two major classes and six types [41]. Type I, III, and IV systems generate the first class of multiprotein complexes. Type I systems use Cas3 endonuclease to cut DNA, while type III Cas10 nuclease cuts RNA. The second class divides into type II, V, and VI systems which comprise a single multi-domain protein with multiple functions within the CRISPR process. Cas9 and Cas12 endonucleases cleave double-stranded DNA (dsDNA) targeted by a crRNA in type II and V, respectively. Cas12 also cuts stranded-single DNA (ssDNA) nonspecifically through a target-dependent activity [27, 42]. Cas13, as a class 2 type VI, can be reprogrammed to cleave the target stranded-single RNA (ssRNA) guided by crRNA, beyond dsDNA. Moreover, Cas13 has a degrading activity for non-target RNA molecules [43]. The corresponding properties of CRISPR types are summarized in Table 1. Class 2 systems are used widely in the CRISPR diagnostics of infectious diseases due to their simplicity and high efficiency [40]. CRISPR/Cas complexes consist of high-sensitivity enzymatic reporters for pathogen detection [44].
In this review, the CRISPR-Dx systems are described, as well as their applications in point-of-care molecular diagnosis of SARS-CoV-2. We have proposed various solutions based on CRISPR-Dx for the challenges of conventional diagnostics. We also go through how CRISPR-Cas systems, including CRISPR-Cas12, CRISPR-Cas13, CRISPR-Cas9, and CRISPR-Cas3, have developed for fast, accurate, and portable diagnostic tests in COVID-19 Pneumonia.
Evidence Acquisition
A literature search has been conducted using PubMed/MEDLINE, ScienceDirect, Scopus, Web of Science, and Google Scholar in pursuit of the proposed solutions by CRISPR-Dx to the challenges of conventional COVID-19 diagnostics. Selected articles in English have been systematically assessed, and the preferred reporting items have been utilized until January 10, 2022.
CRISPR-Based Diagnostics (CRISPR-Dx)
The CRISPR-Dx technology can correctly identify various emerging viruses, such as SARS-CoV-2 mutants. The gRNA can be designed based on conserved regions to recognize the virus even if the genome has mutated. One of these systems is the DNA endonuclease-targeted CRISPR trans-reporter (DETECTR) based on Cas12, which rapidly detects and cleaves ssDNA molecules (Fig. 1) [27]. Recently, nucleic acid pre-amplification has been combined with CRISPR/Cas13 to develop another system called specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) [37]. The SHERLOCK method uses the activity of the Cas13 protein to cleave fluorescent RNA reporters. There is no need for advanced tools and equipment. The DETECTR and SHERLOCK systems are comparable to traditional MDx, such as RT-PCR, in terms of accuracy, specificity, and sensitivity. Both methods are accessible as lateral flow strips. They can also be portable and ultrasensitive. Other advantages of CRISPR-Dx over RT-PCR include the use of isothermal amplification, without specialized thermocycling equipment, high speed to achieve the final result, and specificity to target single nucleotides. Moreover, the CRISPR system can distinguish several pathogens or even various serotypes in the same sample. The SHERLOCK and DETECTR kits have been approved for SARS-CoV-2 detection and are now commercially accessible.
Schematic illustration of DETECTR and SHERLOCK workflows for SARS-CoV-2 detection. DETECTR begins with an RT-PRA step to amplify extracted RNA and then uses the crRNA-Cas12 complex to detect dsDNA targets. On the other hand, SHERLOCK uses T7 transcription to recover RNA amplicons after RT-PRA amplification, and then identifies ssRNA targets utilizing the Cas13-crRNA complex. Cas12 and Cas13 cleave ssDNA and ssRNA reporters, respectively. The results are visible using a lateral flow assay or a fluorescent plate reader. This figure was created by BioRender (http://www.biorender.com)
SARS-CoV-2 CRISPR-Dx
In general, CRISPR-Dx consists of two main elements: (a) a Cas-gRNA (RNP) complex that detects and cuts the specified nucleic acid sequences and (b) nucleotide reporters that create a visual signal when subsequently cut [48]. Different Cas groups have been used for detecting SARS-CoV-2, including Cas3, FnCas9 (Francisella novicida-derived Cas9), Cas12, and Cas13 protein [49]. In particular, the Cas12 protein recognizes and cleaves dsDNA or ssDNA sequences to generate the CRISPR-based SARS-CoV-2 diagnostic [50]. This nuclease is specifically for distinguishing between very similar dsDNA sequences compared to ssDNA. The collateral activity of Cas13 can identify and cleave reporter molecules, identical to Cas12 activity in diagnostic applications [51]. The Cas9 endonuclease is commonly used in genome editing for specific identification and cleavage of DNA sequences through gRNA sequences. However, FnCas9 can be sensitive to single-nucleotide variations of RNA SARS-CoV-2 at temperatures ranging from 10 to 50 °C [52]. Furthermore, like Cas9, the Cas3 endonuclease recognizes DNA sequences and requires a PAM sequence to cut the target site [53].
SARS-CoV-2 CRISPR-Dx Based on Cas12
The first CRISPR/Cas12 DETECTR was combined with reverse transcription and isothermal amplification by loop-mediated replication (RT-LAMP) for RNA SARS-CoV-2 detection [50]. Recently, isothermal amplification methods include recombinase polymerase amplification (RPA) [54] and mediated-loop isothermal amplification (LAMP) [55]. They have more simplicity, rapidity, and low cost compared to PCR. However, non-specific amplification signals and false positives cause a challenge in applying them for accurate and reliable POC testing for clinical diagnostics. Now if only CRISPR/Cas12 combines with RPA pre-amplication, collateral single-stranded DNA can be cleaved surrounding specific gRNA-targeted duplexes [56, 57]. It has been used to develop DETECTR for the detection of the low copy number of SARS-CoV-2 from nasopharyngeal or oropharyngeal swabs in less than 40 min [58]. Cas12 confirms the presence of a virus using cleavage of single-stranded DNA reporters after gRNA targeting (Fig. 1). In contrast to qRT-PCR, DETECTR does not need thermocycling and sophisticated laboratory equipment. The CRISPR/Cas12 DETECTR system has been validated for 36 patients with COVID-19 versus 42 patients with different respiratory viral diseases [59]. This system detected 2-CoV-SARS with 95% positive and 100% negative predictions agreements.
CRISPR/Cas12a-NER is an accelerated diagnostic system that reads the results as green fluorescent light at 458 nm that is visible to the naked eye [60]. When at least ten copies of a viral gene from SARS-CoV-2 are present in the sample, the Cas12 protein cleaves the reporter molecule and detects the results in 40 min. CRISPR/Cas12a-NER is a low-cost, high-throughput, highly sensitive, and effective method to detect even asymptomatic patients [61]. Although most CRISPR-Dx methods use custom paper strips to sense the output signal, there is no need for advanced equipment. The limit of detection (LoD) is reduced compared to fluorescence-based methods. Owing to Poisson sampling distribution, replica variations are significant when template copies are adjusted to be small (below 3–4 copies/Rx) (Fig. 2) [62, 63].
Summary of current CRISPR-Dx technologies organized by Cas12 enzyme to diagnose SARS-CoV-2. The crRNA-Cas12 complex can recognize a specific sequence at the RT-amplified dsDNA. By base pairing between crRNA and target, Cas12 is activated and cleaves the fluorescent ssDNA reporters. Therefore, a fluorescent signal appears as visible light at 458 nm. It can be detected by the naked eye in the CRISPR/Cas12a-NER assay, fluorescent plate reader in the CRISPR-FDS test, or the lateral flow in the iSCAN system. In the AIOD-CRISPR assay, the steps are implemented in a single-pot reaction and incubated at the same temperatures. The SENA test uses rRT-PCR to amplify the SARS-CoV-2 genome and Cas12a to target specific sequences. This figure was created using BioRender (http://www.biorender.com)
A CRISPR-Cas12 detection system has been optimized in 96-well microtiter plates to be used in well-equipped laboratories with precision fluorescent plate readers. This system, named CRISPR-FDS, can be customized with any diagnostic probe designed for RPA or RT-PCR methods (Fig. 2) [64]. It can detect two copies of the target sequence for each positive SARS-CoV-2 sample in about 50 min. For specific samples, the achieved positive results from the sensitive CRISPR-FDS system were false-negative results of the RT-qPCR test. CRISPR-FDS presents a reliable, accurate, and rapid system for COVID-19 detection, although it cannot quantify viral titers like RT-qPCR. In a similar attempt, ultrasensitive detection of SARS-CoV- 2 named iSCAN (in vitro Specific CRISPR-based Assay for Nucleic acids detection) was designed by coupling CRISPR/Cas12a with RT-LAMP [65]. The iSCAN system includes a colorimetric reaction with the lateral immune-chromatography flow to increase the speed, precision, and operation facility (Fig. 2). The iSCAN system is preferred for large-scale, in-field deployment for early identification of SARS-CoV-2 to limit the spread of the virus [66].
In another attempt, an integrated viral nucleic acid detection system called CASdetect (CRISPR-assisted detection) was advanced to detect SARS-CoV-2 [67]. It integrates Cas12b-mediated DNA detection with sample treatment and amplification strategies. The CASdetect system can detect 1 × 104 copies/mL for the SARS-CoV-2 sample, without cross-reactivity to SARS-CoV-1 and MERS-CoV. For more sensitive detection, a CRISPR-Cas12-based specific enhancer was needed, i.e., in the Specific Enhancer for detection of PCR-amplified Nucleic Acids (SENA) [68]. They first analyzed the COVID-19 clinical specimens by rRT-PCR and then verified amplicons with uncertain readouts by SENA. The Cas12a specifically trans-cleaves the rRT-PCR amplicons of the SARS-CoV-2 sequences by mixing fluorescence change (FC) with SENA reaction (mix-FCratio). LoD per reaction was at least two copies less than that of rRT-PCR. The SENA system can detect the virus even in false-negative samples from recovered patients (Fig. 2).
However, multiple manual operations of CRISPR-Dx, such as nucleic acid separation and pre-amplification, complicate the procedures and increase the risk of contamination. An all-in-one dual CRISPR-Cas12a, called “AIOD-CRISPR,” has been developed to resolve this problem for simple, rapid, ultrasensitive, specific, and optic-based detection of SARS-CoV-2. They integrated nucleic acid amplification and a pair of crRNAs for a dual CRISPR-Cas12-based detection in a single reaction (Fig. 2). The AIOD-CRISPR system can detect both retroviral genomes, RNA SARS-CoV-2 and DNA HIV-1, in 20 min. Moreover, a low-cost hand warmer (~ $ 0.3) was used as an incubator for visual detection of COVID-19 at the POC, which has significant potential for developing countries. The AIOD-CRISPR method is the first system in which all components incubate in one reaction without separating manual operations [69]. The AIOD-CRISPR assay enables super-fast (few minutes), ultrasensitive (few copies), and precise detection of SARS-CoV–2 without any false positives (e.g., SARS-CoV, MERS-CoV) with as low as 4.6 copies [70]. Multiplexing rapid detection can be developed by combining the AIOD-CRISPR assay with multiplexed microfluidics technology [71, 72]. Additionally, all reagents can be pre-stored as lyophilized powder in a disposable microfluidic platform [73,74,75,76,77]. With smartphone technology, AIOD-CRISPR signals can be taken as fluorescence photos, converted into fluorescence intensity, analyzed, and reported as qualitative/semi-quantitative test results [78,79,80]. A CRISPR-Cas12a-powered optical biosensor was recently released for ultrasensitive detection of SARS-CoV-2 in a friendly user interface, even with the naked eye. The colorimetric readouts were examined directly on a smartphone running the Color Picker App, substantially simplifying the process, increasing detection mobility, and lowering costs [81]. Overall, a microfluidic device can implement two main steps: (1) automatic nucleic acid extraction from nasopharyngeal samples and healthy controls and (2) an electric field application to simultaneously influence and monitor the activity of the CRISPR/Cas12 complex [82]. Finally, it can detect SARS-CoV-2 nucleic acid in COVID-positive-19 cases in just 30 min.
SARS-CoV-2 CRISPR-Dx Based on Cas13
The CRISPR/Cas13 system is activated by base pairing between the target sequence and crRNA to create a programmable and accurate diagnostic system [56]. An advanced protocol has been established for using the CRISPR/Cas13-based SHERLOCK technique to detect COVID-19 [83]. The test starts with RNA purification from specimens and isothermal RPA amplification. Subsequently, Cas13 detects pre-amplified sequences by destroying supplied reporter RNAs and releasing fluorescent reporters. COVID-19 detection results are read out using dipsticks without requiring elaborate instrumentation. The SHERLOCK COVID-19 detection test ends in 1 h with minimized off-targets by selecting guide sequences in a range between 20 and 200 aM (10–100 copies per microliter of input) [83].
On May 6, 2020, the US Food and Drug Administration (FDA) granted an Emergency Use Authorization (EUA) to Sherlock Biosciences Company favorably considering innovative workflows more divergent than qRT-PCR. It was the first CRISPR-based SARS-CoV-2 test to be approved. Sherlock’s EUA represents a relatively conservative step in a new assay. However, it requires nucleic acid extraction, RPA amplification, a laboratory, trained technicians, and at least one hour or more to run the assays. However, an alternative POC assay named STOPCovid uses a simple lysis buffer to extract the viral genome and a lateral flow dipstick to assess the LoD of 100 copies per reaction in either NP swabs or saliva within 15 to 20 min [84]. New alternatives to SHERLOCK were made using multiplexed nucleic acid detection based on Cas13 called “CARMEN,” which further expands on concepts of microfluidics to screen respiratory pathogens [85]. The enzyme Cas13 from Leptotrichia wadei was used in the clinical validation of the SHERLOCK tests to detect SARS-CoV-2 in samples collected at Siriraj Hospital, Thailand [86]. SHERLOCK was 100% specific and 96% sensitive with a fluorescence readout in 100% agreement with quantitative PCR with reverse transcription [87].
The SHERLOCK technique applied a system called heating unextracted samples to obliterate the nucleases (HUDSON) to eliminate the nucleic acid extraction step (Fig. 3) [88]. This process needs to prepare multiple reactions and transfer samples between them. In a worthy attempt, a fast and sensitive system was made to detect the SARS-CoV-2 genome called “SHINE” (SHERLOCK and HUDSON integration to navigate epidemics) without requiring extraction of nucleic acids [89]. The SHINE system applies portable devices to heat samples with HUDSON in paper-based colorimetric or in-tube fluorescent readout methods with a low chance of contamination. The SHINE is a paper-based colorimetric technique with higher specificity (100%) and sensitivity (90%) compared to RT-qPCR. This method has a sample-to-result period of 50 min (Fig. 3). It was developed for continuous CRISPR-based surveillance using a genomic-comprehensive machine learning design for a series of COVID-19 detection tests and procedures [90]. Developed algorithms improved the sensitivity of SHERLOCK diagnostics for SARS-CoV-2 detection by up to 10 copies per microliter [90].
Summary of current CRISPR-Dx technologies organized by Cas13 enzyme to diagnose SARS-CoV-2. The SHINE test, which combines SHERLOCK and HUDSON, can detect the target sequence without requiring nucleic acid extraction. In the HUDSON step, viral and nuclease are inactivated by heating to prepare for the second step. Without nucleic acid purification, RNA is RT-PRA amplified and recovered using T7 transcription. The crRNA molecule targets a specific sequence, and the Cas13 enzyme cleaves reporters in a single-step SHERLOCK assay. On the other hand, the CREST assay begins with RNA extraction and reverse transcription to DNA molecules. After transcription and targeting, Cas13 cleaves the reporter molecules to detect visible light at 495 nm with a blue LED and an orange filter. This figure was created using BioRender (http://www.biorender.com)
While different CRISPR-Dx systems have been developed for rapid SARS-CoV-2 detection, none have addressed rearrangements and genomic mutations. It is well known that RNA SARS-CoV-2 frequently mutates to avoid attacks from humans’ immunity and constantly adapts. Significantly, such mutations can affect the ability of qRT-PCR assays and the CRISPR/Cas system to recognize the target [91, 92]. Therefore, variant nucleotide guards must be altered to develop the diagnostic system's capabilities for the specific detection of SARS-CoV-2 mutants [93]. By taking advantage of low-cost thermocyclers and widely available enzymes, a method called CREST (Cas13-based, Rugged, Equitable, Scalable Testing) was developed to address the barriers of CRISPR/Cas13-based diagnostics [51]. The CREST technique conveniently detects SARS-CoV-2 using a transcription-recognition reaction based on the PCR method’s reliability and robustness. With management in ∼ 2 h, CREST can be started from standard sampling, high-quality RNA extraction, and reverse transcription to DNA molecules. After amplification, Cas13 activation is visualized with blue LED (~ 495 nm) and orange filters (Fig. 3) [51].
SARS-CoV-2 CRISPR-Dx Based on FnCas9 and Cas3
Among the CRISPR effector enzymes, Cas9 is an RNA-targeted DNase used for gene editing in research settings and gene therapy. Francisella novicida Cas9 (FnCas9) was reported as a very specific nuclease that binds to off-target loci [94]. The FnCas9 system was used to develop an effective and accurate alternative method for diagnosing SARS-CoV-2 called the FnCas9 editor-linked uniform detection assay (FELUDA), which does not require complex instruments [95]. The FELUDA approach utilizes the FnCas9 RNP complex to clarify signatures of the SARS-CoV-2-specific sequences within one hour, with specificity down to one nucleotide for SARS-CoV-1. Such devices prove the efficacy of lateral flow instruments as low-cost and machine-independent alternatives to conventional detection methods.
Recently, it has been reported that Cas3-RNP complexes can trigger specific and targeted cleavage of DNA [96, 97]. A Cas3-based assay was combined with isothermal amplification and was named Cas3-operated nucleic acid detection N (CONAN) [53]. The sensitivity and speed of CONAN RT-LAMP are comparable with those of DETCTR RT-LAMP and qRT-PCR, although it is more specific for single-base-pair discrimination without high-tech performance.
Perspectives
Historically, a broad class of viruses has affected human health, including the H1N1 influenza virus, Human Immunodeficiency Virus (HIV), Zika virus (ZIKV), Ebola Virus, dengue virus (DENV), SARS coronavirus (SARS-CoV), and MERS coronavirus (MERS-CoV). The main challenge in viral outbreaks is rapid testing to limit the spread of the disease [98].
CRISPR-based diagnostic systems have been extensively explored within the field of viral infection in the laboratory and clinical settings [99]. The DETECTR method can distinguish between two high-risk types of human papillomavirus (HPV), HPV16 and HPV18, by targeting the hypervariable loop V of the L1-encoding gene [27]. Furthermore, the SHERLOCK protocol can be used for diagnosing HIV, as one of the significant concerns worldwide [32, 88]. A PCR-based CRISPR-Cas13a detects the drug resistance mutation in the hepatitis B virus (HBV) [100]. SHERLOCK and HUDSON protocols can also diagnose flaviviruses such as ZIKV, DENV, West Nile, and yellow fever viruses with high sensitivity and single-base resolution [101]. In addition, several other viruses were also detected using CRISPR-Cas12a/based-Cas13a methods, such as avian influenza A (H7N9) viruses [102], Ebola virus (EBOV), and Lassa virus (LASV) [103, 104], as well as SARS-CoV-2 [105, 106].
CRISPR proteins play a crucial role in bacterial and archaeal immune defense by allowing the identification and cleavage of specific foreign nucleic acid sequences, including invading viruses or plasmids [107, 108]. The CRISPR-Dx system has been recognized as a practical tool in identifying antimicrobial drug-resistant bacteria [109]. One method called FLASH (Finding Low Abundance Sequences by Hybridization) has used Cas9 and multiplex guide RNAs to target a total of 3624 antimicrobial resistant bacteria, including S. aureus, MRSA infections, and vancomycin-resistant E. faecium [110]. Besides FLASH, the CRISPR-Cas12a system has usability for rapid Mycobacterium tuberculosis (Mtb) diagnosis following RPA [111]. During the enterohemorrhagic outbreak in 2011, CRISPR techniques could detect a specific locus to the E. coli hybrid [112]. The CRISPR-Dx system plays an important role in determining the oncogenes and genetic mechanisms in cancer research. Post-treatment changes and resistance genes to drugs can be targeted to identify new accuracy therapy biomarkers and cancer prevention [113, 114]. The CRISPR-based systems recognize the desired genomic area via a high-specific gRNA and make a break using one of the Cas nucleases. They can be reprogrammed to ensure that unwanted products do not interfere with the reaction outcome, even in the presence of similar mutates.
Conclusions
There are currently more than 531 million confirmed COVID-19 cases worldwide and 6.3 million confirmed deaths, according to the last update from WHO (May 30, 2022, 08:16 GMT). Effective screening methods have a critical role in controlling the pandemic. Now, RT-PCR provides adequate sensitivity and specificity. However, it lacks rapidity and cost-effectiveness. The results of RT-PCR tests are determined in 3–4 h, and an average of 6–8 h is added to this time because samples must be sent to specialized laboratories. Additionally, the high-tech equipment and trained operators essential for this method are significant limitations to its use. Serological screening approaches show lower sensitivity and specificity due to cross-reactivity with the phylogenetically closest viruses, despite of rapidity and affordability. Also, immunoassays cannot detect SARS-CoV-2-specific antibodies during the early stages of COVID-19, leading to false-negative serological results. Moreover, there would be individual variability in the host immune response, representing population-level allelic diversity of antibodies. Antigen tests also detect viral surface proteins to diagnose an active or acute infection in specimens from the nasopharynx and anterior nares. It is recommended antigen tests for individuals, with symptoms during the first 5 to 7 days of infection with an average sensitivity of about 50%. Thus, there is an urgent need to control the spread of disease through instrument-free point-of-care detections that are specific, sensitive, low cost, and rapidly field-deployable all at once. Such devices are the primary goal of CRISPR-based systems. They will ambitiously compete for diagnostics and possibly have a lasting and effective impact on best practices and state-of-the-art methods in the post-pandemic era. To conclude, CRISPR-based systems can be easily used even in communities with poor economies since no sophisticated laboratory equipment is needed.
References
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., & Huang, C.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273.
Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., & Pei, Y.-Y. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579, 265–269.
Cui, J., Li, F., & Shi, Z.-L. (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology, 17, 181–192.
Frutos, R., Serra-Cobo, J., Pinault, L., Lopez Roig, M., & Devaux, C. A. (2021). Emergence of bat-related betacoronaviruses: Hazard and risks. Frontiers in Microbiology, 12, 437.
World Health Organization. (2021). Genomic sequencing of SARS-CoV-2: A guide to implementation for maximum impact on public health. World Health Organization.
Vieira, C. M., Franco, O. H., Restrepo, C. G., & Abel, T. (2020). COVID-19: The forgotten priorities of the pandemic. Maturitas, 136, 38–41.
Sood, S., Aggarwal, V., Aggarwal, D., Upadhyay, S. K., Sak, K., Tuli, H. S., Kumar, M., Kumar, J., & Talwar, S. (2020). COVID-19 pandemic: From molecular biology, pathogenesis, detection, and treatment to global societal impact. Current Pharmacology Reports, 6, 1–16.
Sun, K., Wang, W., Gao, L., Wang, Y., Luo, K., Ren, L., Zhan, Z., Chen, X., Zhao, S., & Huang, Y. (2021). Transmission heterogeneities, kinetics, and controllability of SARS-CoV-2. Science. https://doi.org/10.1126/science.abe2424
Han, Y., & Yang, H. (2020). The transmission and diagnosis of 2019 novel coronavirus infection disease (COVID-19): A Chinese perspective. Journal of Medical Virology, 92, 639–644.
Döhla, M., Boesecke, C., Schulte, B., Diegmann, C., Sib, E., Richter, E., Eschbach-Bludau, M., Aldabbagh, S., Marx, B., & Eis-Hübinger, A.-M. (2020). Rapid point-of-care testing for SARS-CoV-2 in a community screening setting shows low sensitivity. Public Health, 182, 170–172.
Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K., Bleicker, T., Brünink, S., Schneider, J., & Schmidt, M. L. (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance, 25, 2000045.
Covid, C., Team, R., COVID, C., Team, R., COVID, C., Team, R., Burrer, S. L., de Perio, M. A., Hughes, M. M., & Kuhar, D. T. (2020). Characteristics of health care personnel with COVID-19—United States, February 12–April 9, 2020. Morbidity and Mortality Weekly Report, 69, 477.
World Health Organization. (2020). Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: Interim guidance, 19 March 2020. World Health Organization.
Morshed, M. G., Lee, M.-K., Jorgensen, D., & Isaac-Renton, J. L. (2007). Molecular methods used in clinical laboratory: Prospects and pitfalls. FEMS Immunology and Medical Microbiology, 49, 184–191.
Nguyen, T., Duong Bang, D., & Wolff, A. (2020). 2019 novel coronavirus disease (COVID-19): Paving the road for rapid detection and point-of-care diagnostics. Micromachines, 11, 306.
Xiao, A. T., Tong, Y. X., & Zhang, S. (2020). False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. Journal of Medical Virology, 92, 1755–1756.
Feng, H., Liu, Y., Lv, M., & Zhong, J. (2020). A case report of COVID-19 with false negative RT-PCR test: Necessity of chest CT. Japanese Journal of Radiology, 38, 409–410.
Li, Y., Yao, L., Li, J., Chen, L., Song, Y., Cai, Z., & Yang, C. (2020). Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. Journal of Medical Virology, 92, 903–908.
Pan, Y., Long, L., Zhang, D., Yuan, T., Cui, S., Yang, P., Wang, Q., & Ren, S. (2020). Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clinical Chemistry, 66, 794–801.
Surkova, E., Nikolayevskyy, V., & Drobniewski, F. (2020). False-positive COVID-19 results: Hidden problems and costs. The Lancet Respiratory Medicine, 8, 1167–1168.
Ramdas, K., Darzi, A., & Jain, S. (2020). ‘Test, re-test, re-test’: Using inaccurate tests to greatly increase the accuracy of COVID-19 testing. Nature Medicine, 26, 810–811.
Tahamtan, A., & Ardebili, A. (2020). Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Review of Molecular Diagnostics, 20, 453–454.
Mabey, D., Peeling, R. W., Ustianowski, A., & Perkins, M. D. (2004). Diagnostics for the developing world. Nature Reviews Microbiology, 2, 231–240.
Mori, Y., & Notomi, T. (2009). Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. Journal of Infection and Chemotherapy, 15, 62–69.
Nagai, K., Horita, N., Yamamoto, M., Tsukahara, T., Nagakura, H., Tashiro, K., Shibata, Y., Watanabe, H., Nakashima, K., & Ushio, R. (2016). Diagnostic test accuracy of loop-mediated isothermal amplification assay for Mycobacterium tuberculosis: Systematic review and meta-analysis. Science and Reports, 6, 1–10.
Li, S.-Y., Cheng, Q.-X., Liu, J.-K., Nie, X.-Q., Zhao, G.-P., & Wang, J. (2018). CRISPR-Cas12a has both cis-and trans-cleavage activities on single-stranded DNA. Cell Research, 28, 491–493.
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., & Doudna, J. A. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 360, 436–439.
Harrington, L. B., Burstein, D., Chen, J. S., Paez-Espino, D., Ma, E., Witte, I. P., Cofsky, J. C., Kyrpides, N. C., Banfield, J. F., & Doudna, J. A. (2018). Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science, 362, 839–842.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821.
Chertow, D. S. (2018). Next-generation diagnostics with CRISPR. Science, 360, 381–382.
Li, S.-Y., Cheng, Q.-X., Wang, J.-M., Li, X.-Y., Zhang, Z.-L., Gao, S., Cao, R.-B., Zhao, G.-P., & Wang, J. (2018). CRISPR-Cas12a-assisted nucleic acid detection. Cell Discovery, 4, 1–4.
Bhattacharyya, R. P., Thakku, S. G., & Hung, D. T. (2018). Harnessing CRISPR effectors for infectious disease diagnostics. ACS Infectious Diseases, 4, 1278–1282.
Batista, A. C., & Pacheco, L. G. (2018). Detecting pathogens with Zinc-Finger, TALE and CRISPR-based programmable nucleic acid binding proteins. Journal of Microbiological Methods, 152, 98–104.
Li, Y., Li, S., Wang, J., & Liu, G. (2019). CRISPR/Cas systems towards next-generation biosensing. Trends in Biotechnology, 37, 730–743.
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., Verdine, V., Cox, D. B., Kellner, M. J., & Regev, A. (2017). RNA targeting with CRISPR–Cas13. Nature, 550, 280–284.
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., Volz, S. E., Joung, J., Van Der Oost, J., & Regev, A. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163, 759–771.
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., & Zhang, F. (2019). SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nature Protocols, 14, 2986–3012.
Gasiunas, G., Barrangou, R., Horvath, P., & Siksnys, V. (2012). Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences, 109, E2579–E2586.
Slaymaker, I. M., Mesa, P., Kellner, M. J., Kannan, S., Brignole, E., Koob, J., Feliciano, P. R., Stella, S., Abudayyeh, O. O., & Gootenberg, J. S. (2019). High-resolution structure of Cas13b and biochemical characterization of RNA targeting and cleavage. Cell Reports, 26(3741–3751), e3745.
van Dongen, J. E., Berendsen, J. T., Steenbergen, R. D., Wolthuis, R. M., Eijkel, J. C., & Segerink, L. I. (2020). Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosensors and Bioelectronics, 166, 112445.
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J., Charpentier, E., Cheng, D., Haft, D. H., & Horvath, P. (2020). Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nature Reviews Microbiology, 18, 67–83.
Chen, Y., Mei, Y., Zhao, X., & Jiang, X. (2020). Reagents-loaded, automated assay that integrates recombinase-aided amplification and Cas12a nucleic acid detection for a point-of-care test. Analytical Chemistry, 92, 14846–14852.
Jia, F., Li, X., Zhang, C., & Tang, X. (2020). The expanded development and application of CRISPR system for sensitive nucleotide detection. Protein & Cell, 11, 624–629.
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., & Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science, 360, 439–444.
Chaudhary, K., Chattopadhyay, A., & Pratap, D. (2018). The evolution of CRISPR/Cas9 and their cousins: Hope or hype? Biotechnology letters, 40, 465–477.
Lee, J. (2020). Anti-CRISPR proteins: Applications in genome engineering. FEBS Journal, 287(4), 626–644.
Garcia-Doval, C., & Jinek, M. (2017). Molecular architectures and mechanisms of Class 2 CRISPR-associated nucleases. Current Opinion in Structural Biology, 47, 157–166.
Shi, K., Xie, S., Tian, R., Wang, S., Lu, Q., Gao, D., Lei, C., Zhu, H., & Nie, Z. (2021). A CRISPR-Cas autocatalysis-driven feedback amplification network for supersensitive DNA diagnostics. Science Advances, 7, 7802.
Srivastava, A., Gupta, T., Kumar, S., & Saxena, S. K. (2020). Next-generation rapid advanced molecular diagnostics of COVID-19 by CRISPR-Cas. Diagnostic strategies for COVID-19 and other coronaviruses (pp. 175–187). Springer.
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Servellita, V., Singh, J., Miao, X., Streithorst, J. A., Granados, A., & Sotomayor-Gonzalez, A. (2020). CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology, 38, 870–874.
Rauch, J. N., Valois, E., Solley, S. C., Braig, F., Lach, R. S., Audouard, M., Ponce-Rojas, J. C., Costello, M. S., Baxter, N. J., & Kosik, K. S. (2021). A scalable, easy-to-deploy protocol for Cas13-based detection of SARS-CoV-2 genetic material. Journal of Clinical Microbiology, 59, e02402-20.
Azhar, M., Phutela, R., Kumar, M., Ansari, A. H., Rauthan, R., Gulati, S., Sharma, N., Sinha, D., Sharma, S., & Singh, S. (2021). Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosensors and Bioelectronics, 183, 113207.
Yoshimi, K., Takeshita, K., Yamayoshi, S., Shibumura, S., Yamauchi, Y., Yamamoto, M., Yotsuyanagi, H., Kawaoka, Y. & Mashimo, T. (2020). Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3.
Piepenburg, O., Williams, C. H., Stemple, D. L., & Armes, N. A. (2006). DNA detection using recombination proteins. PLoS Biology, 4, e204.
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., & Hase, T. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Research, 28, e63–e63.
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B., Shmakov, S., Makarova, K. S., Semenova, E., & Minakhin, L. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science, 353, aaf5573.
Jeon, Y., Choi, Y. H., Jang, Y., Yu, J., Goo, J., Lee, G., Jeong, Y. K., Lee, S. H., Kim, I.-S., & Kim, J.-S. (2018). Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nature Communications, 9, 1–11.
Brandsma, E., Verhagen, H. J., van de Laar, T. J., Claas, E. C., Cornelissen, M. & van den Akker, E. (2020). Rapid, sensitive and specific SARS coronavirus-2 detection: a multi-center comparison between standard qRT-PCR and CRISPR based DETECTR. medRxiv.
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Singh, J., Streithorst, J., Granados, A., Sotomayor-Gonzalez, A., Zorn, K. & Gopez, A. (2020). Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. MedRxiv.
Wang, X., Zhong, M., Liu, Y., Ma, P., Dang, L., Meng, Q., Wan, W., Ma, X., Liu, J., & Yang, G. (2020). Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Science Bulletin, 65(17), 1436–2143.
Zhan, Y., Li, X.-P., & Yin, J.-Y. (2021). COVID-19 one year later: A retrospect of CRISPR-Cas system in combating COVID-19. International Journal of Biological Sciences, 17, 2080–2088.
Burns, M., & Valdivia, H. (2008). Modelling the limit of detection in real-time quantitative PCR. European Food Research and Technology, 226, 1513–1524.
Forootan, A., Sjöback, R., Björkman, J., Sjögreen, B., Linz, L., & Kubista, M. (2017). Methods to determine the limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomolecular Detection and Quantification, 12, 1–6.
Huang, Z., Tian, D., Liu, Y., Lin, Z., Lyon, C. J., Lai, W., Fusco, D., Drouin, A., Yin, X., & Hu, T. (2020). Ultra-sensitive and high-throughput CRISPR-p powered COVID-19 diagnosis. Biosensors & Bioelectronics, 164, 112316.
Ali, Z., Aman, R., Mahas, A., Rao, G. S., Tehseen, M., Marsic, T., Salunke, R., Subudhi, A. K., Hala, S. M., & Hamdan, S. M. (2020). iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Research, 288, 198129.
Palaz, F., Kalkan, A. K., Tozluyurt, A., & Ozsoz, M. (2021). CRISPR-based tools: Alternative methods for the diagnosis of COVID-19. Clinical Biochemistry, 89, 1.
Guo, L., Sun, X., Wang, X., Liang, C., Jiang, H., Gao, Q., Dai, M., Qu, B., Fang, S., & Mao, Y. (2020). SARS-CoV-2 detection with CRISPR diagnostics. Cell Discovery, 6, 1–4.
Huang, W., Yu, L., Wen, D., Wei, D., Sun, Y., Zhao, H., Ye, Y., Chen, W., Zhu, Y., & Wang, L. (2020). A CRISPR-Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. eBioMedicine, 61, 103036.
Wang, B., Wang, R., Wang, D., Wu, J., Li, J., Wang, J., Liu, H., & Wang, Y. (2019). Cas12aVDet: A CRISPR/Cas12a-based platform for rapid and visual nucleic acid detection. Analytical Chemistry, 91, 12156–12161.
Kim, H., Lee, W.-J., Oh, Y., Kang, S.-H., Hur, J. K., Lee, H., Song, W., Lim, K.-S., Park, Y.-H., & Song, B.-S. (2020). Enhancement of target specificity of CRISPR–Cas12a by using a chimeric DNA–RNA guide. Nucleic Acids Research, 48, 8601–8616.
Fang, X., Chen, H., Yu, S., Jiang, X., & Kong, J. (2011). Predicting viruses accurately by a multiplex microfluidic loop-mediated isothermal amplification chip. Analytical Chemistry, 83, 690–695.
Dou, M., Sanjay, S. T., Dominguez, D. C., Liu, P., Xu, F., & Li, X. (2017). Multiplexed instrument-free meningitis diagnosis on a polymer/paper hybrid microfluidic biochip. Biosensors & Bioelectronics, 87, 865–873.
Song, J., Mauk, M. G., Hackett, B. A., Cherry, S., Bau, H. H., & Liu, C. (2016). Instrument-free point-of-care molecular detection of Zika virus. Analytical Chemistry, 88, 7289–7294.
Chen, D., Mauk, M., Qiu, X., Liu, C., Kim, J., Ramprasad, S., Ongagna, S., Abrams, W. R., Malamud, D., & Corstjens, P. L. (2010). An integrated, self-contained microfluidic cassette for isolation, amplification, and detection of nucleic acids. Biomedical Microdevices, 12, 705–719.
Park, S., Zhang, Y., Lin, S., Wang, T.-H., & Yang, S. (2011). Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnology Advances, 29, 830–839.
Ferguson, B. S., Buchsbaum, S. F., Wu, T.-T., Hsieh, K., Xiao, Y., Sun, R., & Soh, H. T. (2011). Genetic analysis of H1N1 influenza virus from throat swab samples in a microfluidic system for point-of-care diagnostics. Journal of the American Chemical Society, 133, 9129–9135.
Song, J., Liu, C., Mauk, M. G., Peng, J., Schoenfeld, T., & Bau, H. H. (2018). A multifunctional reactor with dry-stored reagents for enzymatic amplification of nucleic acids. Analytical Chemistry, 90, 1209–1216.
Chen, W., Yu, H., Sun, F., Ornob, A., Brisbin, R., Ganguli, A., Vemuri, V., Strzebonski, P., Cui, G., & Allen, K. J. (2017). Mobile platform for multiplexed detection and differentiation of disease-specific nucleic acid sequences, using microfluidic loop-mediated isothermal amplification and smartphone detection. Analytical Chemistry, 89, 11219–11226.
Song, J., Pandian, V., Mauk, M. G., Bau, H. H., Cherry, S., Tisi, L. C., & Liu, C. (2018). Smartphone-based mobile detection platform for molecular diagnostics and spatiotemporal disease mapping. Analytical Chemistry, 90, 4823–4831.
Yin, K., Pandian, V., Kadimisetty, K., Ruiz, C., Cooper, K., You, J., & Liu, C. (2019). Synergistically enhanced colorimetric molecular detection using smart cup: A case for instrument-free HPV-associated cancer screening. Theranostics, 9, 2637.
Ma, L., Yin, L., Li, X., Chen, S., Peng, L., Liu, G., Ye, S., Zhang, W., & Man, S. (2022). A smartphone-based visual biosensor for CRISPR-Cas powered SARS-CoV-2 diagnostics. Biosensors and Bioelectronics, 195, 113646.
Ramachandran, A., Huyke, D. A., Sharma, E., Sahoo, M. K., Huang, C., Banaei, N., Pinsky, B. A., & Santiago, J. G. (2020). Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proceedings of the National Academy of Sciences, 117, 29518–29525.
Zhang, F., Abudayyeh, O. O. & Gootenberg, J. S. (2020). A protocol for detection of COVID-19 using CRISPR diagnostics. A protocol for detection of COVID-19 using CRISPR diagnostics 8.
Joung, J., Ladha, A., Saito, M., Segel, M., Bruneau, R., Huang, M.-l. W., Kim, N.-G., Yu, X., Li, J. & Walker, B. D. (2020). Point-of-care testing for COVID-19 using SHERLOCK diagnostics. MedRxiv.
Ackerman, C. M., Myhrvold, C., Thakku, S. G., Freije, C. A., Metsky, H. C., Yang, D. K., Simon, H. Y., Boehm, C. K., Kosoko-Thoroddsen, T.-S.F., & Kehe, J. (2020). Massively multiplexed nucleic acid detection with Cas13. Nature, 582, 277–282.
Patchsung, M., Jantarug, K., Pattama, A., Aphicho, K., Suraritdechachai, S., Meesawat, P., Sappakhaw, K., Leelahakorn, N., Ruenkam, T., & Wongsatit, T. (2020). Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nature Biomedical Engineering, 4, 1140–1149.
Mustafa, M. I., & Makhawi, A. M. (2021). SHERLOCK and DETECTR: CRISPR-Cas systems as potential rapid diagnostic tools for emerging infectious diseases. Journal of Clinical Microbiology. https://doi.org/10.1128/JCM.00745-20
Myhrvold, C., Freije, C. A., Gootenberg, J. S., Abudayyeh, O. O., Metsky, H. C., Durbin, A. F., Kellner, M. J., Tan, A. L., Paul, L. M., & Parham, L. A. (2018). Field-deployable viral diagnostics using CRISPR-Cas13. Science, 360, 444–448.
Arizti-Sanz, J., Freije, C. A., Stanton, A. C., Petros, B. A., Boehm, C. K., Siddiqui, S., Shaw, B. M., Adams, G., Kosoko-Thoroddsen, T.-S.F., & Kemball, M. E. (2020). Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nature Communications, 11, 1–9.
Metsky, H. C., Freije, C. A., Kosoko-Thoroddsen, T.-S. F., Sabeti, P. C. & Myhrvold, C. (2020). CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. BioRxiv.
Wang, R., Hozumi, Y., Yin, C., & Wei, G.-W. (2020). Mutations on COVID-19 diagnostic targets. Genomics, 112, 5204–5213.
van Dorp, L., Acman, M., Richard, D., Shaw, L. P., Ford, C. E., Ormond, L., Owen, C. J., Pang, J., Tan, C. C., & Boshier, F. A. (2020). Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infection, Genetics and Evolution, 83, 104351.
Ooi, K. H., Tay, J. W. D., Teo, S. Y., Liu, M. M., Kaewsapsak, P., Jin, S., Gao, Y.-G. & Tan, M. H. (2020). A CRISPR-based SARS-CoV-2 diagnostic assay that is robust against viral evolution and RNA editing. bioRxiv.
Acharya, S., Mishra, A., Paul, D., Ansari, A. H., Azhar, M., Kumar, M., Rauthan, R., Sharma, N., Aich, M., & Sinha, D. (2019). Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing. Proceedings of the National Academy of Sciences, 116, 20959–20968.
Azhar, M., Phutela, R., Ansari, A. H., Sinha, D., Sharma, N., Kumar, M., Aich, M., Sharma, S., Singhal, K., & Lad, H. (2020). Rapid, field-deployable nucleobase detection and identification using FnCas9. BioRxiv., 360, 381LP.
Morisaka, H., Yoshimi, K., Okuzaki, Y., Gee, P., Kunihiro, Y., Sonpho, E., Xu, H., Sasakawa, N., Naito, Y., & Nakada, S. (2019). CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nature Communications, 10, 1–13.
Dolan, A. E., Hou, Z., Xiao, Y., Gramelspacher, M. J., Heo, J., Howden, S. E., Freddolino, P. L., Ke, A., & Zhang, Y. (2019). Introducing a spectrum of long-range genomic deletions in human embryonic stem cells using type I CRISPR-Cas. Molecular Cell, 74(936–950), e935.
Sridhara, S., Goswami, H. N., Whyms, C., Dennis, J. H., & Li, H. (2021). Virus detection via programmable Type III-A CRISPR-Cas systems. Nature Communications, 12, 1–10.
Kocak, D. D., & Gersbach, C. A. (2018). From CRISPR scissors to virus sensors. Nature, 557, 168.
Wang, S., Li, H., Kou, Z., Ren, F., Jin, Y., Yang, L., Dong, X., Yang, M., Zhao, J., & Liu, H. (2021). Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clinical Microbiology and Infection, 27, 443–450.
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., Verdine, V., Donghia, N., Daringer, N. M., & Freije, C. A. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science, 356, 438–442.
Liu, Y., Xu, H., Liu, C., Peng, L., Khan, H., Cui, L., Huang, R., Wu, C., Shen, S., & Wang, S. (2019). CRISPR-Cas13a nanomachine based simple technology for avian influenza A (H7N9) virus on-site detection. Journal of Biomedical Nanotechnology, 15, 790–798.
Barnes, K. G., Lachenauer, A. E., Nitido, A., Siddiqui, S., Gross, R., Beitzel, B., Siddle, K. J., Freije, C. A., Dighero-Kemp, B., & Mehta, S. B. (2020). Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real time. Nature Communications, 11, 1–10.
Qin, P., Park, M., Alfson, K. J., Tamhankar, M., Carrion, R., Patterson, J. L., Griffiths, A., He, Q., Yildiz, A., & Mathies, R. (2019). Rapid and fully microfluidic Ebola virus detection with CRISPR-Cas13a. ACS Sensors, 4, 1048–1054.
Hou, T., Zeng, W., Yang, M., Chen, W., Ren, L., Ai, J., Wu, J., Liao, Y., Gou, X., & Li, Y. (2020). Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathogens, 16, e1008705.
Fozouni, P., Son, S., de León Derby, M. D., Knott, G. J., Gray, C. N., D’Ambrosio, M. V., Zhao, C., Switz, N. A., Kumar, G. R., & Stephens, S. I. (2021). Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell, 184(323–333), e329.
Shmakov, S., Smargon, A., Scott, D., Cox, D., Pyzocha, N., Yan, W., Abudayyeh, O. O., Gootenberg, J. S., Makarova, K. S., & Wolf, Y. I. (2017). Diversity and evolution of class 2 CRISPR–Cas systems. Nature Reviews Microbiology, 15, 169–182.
Marraffini, L. A. (2015). CRISPR-Cas immunity in prokaryotes. Nature, 526, 55–61.
Wang, W., Hou, J., Zheng, N., Wang, X., & Zhang, J. (2019). Keeping our eyes on CRISPR: The “Atlas” of gene editing (Vol. 35, pp. 285–288). New York: Springer.
Quan, J., Langelier, C., Kuchta, A., Batson, J., Teyssier, N., Lyden, A., Caldera, S., McGeever, A., Dimitrov, B., & King, R. (2019). FLASH: A next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Research, 47, e83–e83.
Ai, J.-W., Zhou, X., Xu, T., Yang, M., Chen, Y., He, G.-Q., Pan, N., Cai, Y., Li, Y., & Wang, X. (2019). CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerging Microbes & Infections, 8, 1361–1369.
Delannoy, S., Beutin, L., Burgos, Y., & Fach, P. (2012). Specific detection of enteroaggregative hemorrhagic Escherichia coli O104: H4 strains by use of the CRISPR locus as a target for a diagnostic real-time PCR. Journal of Clinical Microbiology, 50, 3485–3492.
Tian, X., Gu, T., Patel, S., Bode, A. M., Lee, M.-H., & Dong, Z. (2019). CRISPR/Cas9–An evolving biological tool kit for cancer biology and oncology. NPJ Precision Oncology, 3, 1–8.
Khambhati, K., Bhattacharjee, G., & Singh, V. (2019). Current progress in CRISPR-based diagnostic platforms. Journal of Cellular Biochemistry, 120, 2721–2725.
Funding
No external funding was used in the preparation of this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nafian, F., Nafian, S., Kamali Doust Azad, B. et al. CRISPR-Based Diagnostics and Microfluidics for COVID-19 Point-of-Care Testing: A Review of Main Applications. Mol Biotechnol 65, 497–508 (2023). https://doi.org/10.1007/s12033-022-00570-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00570-5