Abstract
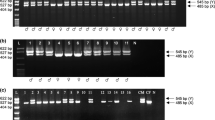
Sex selection technologies have immensely impacted swine production globally. Conventional earlier embryo sex identification methods require professional technicians and sophisticated laboratory instruments. Rapid on-site gender identification of porcine embryos and pork products remains challenging. In this study, we developed a CRISPR/Cas12a-based fluorescence visualization point-of-care sex determination test that is rapid, accurate and easy to implement on-site. The CRISPR/Cas12a assay coupled with either the polymerase chain reaction (PCR) or loop-mediated isothermal amplification (LAMP) employs precisely designed primers and single-guide RNAs targeting the sex-determining region Y (SRY) and the zinc finger protein X-linked (ZFX) genes. PCR and LAMP amplicons were cleaved with the subsequent generation of fluorescing products detectable with portable blue and ultraviolet light transilluminators. Approximately two copies per microliter of the ZFX and SRY genes were detected using the RApid VIsual CRISPR (RAVI-CRISPR) assay. This method is a sensitive, inexpensive, versatile, and point-of-care test. The technology has other potential applications like determining the sex of diverse livestock species, detecting livestock disease-causing pathogens and evaluating the quality of meat products.






Similar content being viewed by others
References
Osada, M., Iwabuchi, H., Aoki, T., Sasaki, K., Ushijima, H., & Ozawa, T. (2019). Economic evaluation of artificial insemination of sex-sorted semen on a Brown Swiss dairy farm-A case study. Animal Science Journal, 90(4), 597–603. https://doi.org/10.1111/asj.13156
Seidel Jr, G. E. (2014). Update on sexed semen technology in cattle. Animal, 8(Suppl 1), 160–164. https://doi.org/10.1017/S1751731114000202
Schinckel, A. P., Mahan, D. C., Wiseman, T. G., & Einstein, M. E. (2008). Growth of protein, moisture, lipid, and ash of two genetic lines of barrows and gilts from twenty to one hundred twenty-five kilograms of body weight. Journal of Animal Science, 86(2), 460–471. https://doi.org/10.2527/jas.2007-0625
Gálvez, F., Domínguez, R., Pateiro, M., Carballo, J., Tomasevic, I., & Lorenzo, J. M. (2018). Effect of gender on breast and thigh turkey meat quality. British Poultry Science, 59(4), 408–415. https://doi.org/10.1080/00071668.2018.1465177
Carabús, A., Sainz, R. D., Oltjen, J. W., Gispert, M., & Font-I-Furnols, M. (2017). Growth of total fat and lean and of primal cuts is affected by the sex type. Animal, 11(8), 1321–1329. https://doi.org/10.1017/S1751731117000039
Kim, Y. M., Choi, T. J., Ho Cho, K., Cho, E. S., Lee, J. J., Chung, H. J., Baek, S. Y., & Jeong, Y. D. (2018). Effects of sex and breed on meat quality and sensory properties in three-way crossbred pigs sired by duroc or by a synthetic breed based on a korean native breed. Korean Journal for Food Science of Animal Resources, 38(3), 544–553. https://doi.org/10.5851/kosfa.2018.38.3.544
Claus, R., Weiler, U., & Herzog, A. (1994). Physiological aspects of androstenone and skatole formation in the boar-A review with experimental data. Meat Science, 38(2), 289–305. https://doi.org/10.1016/0309-1740(94)90118-X
Gunawan, A., Sahadevan, S., Neuhoff, C., Große-Brinkhaus, C., Gad, A., Frieden, L., Tesfaye, D., Tholen, E., Looft, C., Uddin, M. J., Schellander, K., & Cinar, M. U. (2013). RNA deep sequencing reveals novel candidate genes and polymorphisms in boar testis and liver tissues with divergent androstenone levels. PLoS ONE, 8(5), e63259. https://doi.org/10.1371/journal.pone.0063259
Bañón, S., Andreu, C., Laencina, J., & Garro ida.-D, M. (2004). Fresh and eating pork quality from entire versus castrate heavy males. Food Quality and Preference, 15(3), 293–300. https://doi.org/10.1016/S0950-3293(03)00069-7
Lee, G. J., Archibald, A. L., Law, A. S., Lloyd, S., Wood, J., & Haley, C. S. (2005). Detection of quantitative trait loci for androstenone, skatole and boar taint in a cross between Large White and Meishan pigs. Animal Genetics, 36(1), 14–22. https://doi.org/10.1111/j.1365-2052.2004.01214.x
Espinosa-Cervantes, R., & Córdova-Izquierdo, A. (2013). Sexing sperm of domestic animals. Tropical Animal Health and Production, 45(1), 1–8. https://doi.org/10.1007/s11250-012-0215-0
Polge, C. (1985). How does embryo manipulation fit into present and future pig reproduction? Journal of Reproduction and Fertility. Supplement, 33, 93–100.
Garner, D. L., Evans, K. M., & Seidel, G. E. (2013). Sex-sorting sperm using flow cytometry/cell sorting Methods in Molecular Biology (Clifton, N.J.). Humana Press, 927, 279–295.
Rath, D., & Johnson, L. A. (2008). Application and commercialization of flow cytometrically sex-sorted semen. Reproduction in Domestic Animals, 43(Suppl 2), 338–346. https://doi.org/10.1111/j.1439-0531.2008.01182.x
Seok, S. H., Kang, S. Y., Im, Y. B., Yoo, H. S., & Yeon, S. C. (2019). Sex identification using ZFX and ZFY genes in leopard cats (Prionailurus bengalensis euptilurus) in Korea. Journal of Veterinary Medical Science, 81(5), 793–798. https://doi.org/10.1292/jvms.18-0693
Pomp, D., Good, B. A., Geisert, R. D., Corbin, C. J., & Conley, A. J. (1995). Sex identification in mammals with polymerase chain reaction and its use to examine sex effects on diameter of day-10 or -11 pig embryos. Journal of Animal Science, 73(5), 1408–1415. https://doi.org/10.2527/1995.7351408x
Peura, T., Hyttinen, J. M., Turunen, M., & Jänne, J. (1991). Areliable sex determination assay for bovine preimplantation embryos using the polymerase chain reaction. Theriogenology, 35(3), 547–555. https://doi.org/10.1016/0093-691x(91)90451-i
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., & Hase, T. (2000). Loop-mediated isothermal amplification of DNA. Nucleic acids research, 28(12), E63. https://doi.org/10.1093/nar/28.12.e63
Almasi, M. A., & Almasi, G. (2017). Loop mediated isothermal amplification (lamp) for embryo sex determination in pregnant women at eight weeks of pregnancy. Journal of Reproduction & Infertility, 18(1), 197–204.
Zoheir, K. M., & Allam, A. A. (2010). A rapid method for sexing the bovine embryo. Animal Reproduction Science, 119(1–2), 92–96. https://doi.org/10.1016/j.anireprosci.2009.12.013
Hirayama, H., Kageyama, S., Takahashi, Y., Moriyasu, S., Sawai, K., Onoe, S., Watanabe, K., Kojiya, S., Notomi, T., & Minamihashi, A. (2006). Rapid sexing of water buffalo (Bubalus bubalis) embryos using loop-mediated isothermal amplification. Theriogenology, 66(5), 1249–1256. https://doi.org/10.1016/j.theriogenology.2006.03.036
Wang, Y., Liu, D., Deng, J., Wang, Y., Xu, J., & Ye, C. (2017). Loop-mediated isothermal amplification using self-avoiding molecular recognition systems and antarctic thermal sensitive uracil-DNA-glycosylase for detection of nucleic acid with prevention of carryover contamination. Analytica Chimica Acta, 996, 74–87. https://doi.org/10.1016/j.aca.2017.10.022
Borst, A., Box, A. T., & Fluit, A. C. (2004). False-positive results and contamination in nucleic acid amplification assays: Suggestions for a prevent and destroy strategy. European Journal of Clinical Microbiology & Infectious Diseases, 23(4), 289–299. https://doi.org/10.1007/s10096-004-1100-1
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., & Doudna, J. A. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 360(6387), 436–439. https://doi.org/10.1126/science.aar6245
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., Verdine, V., Donghia, N., Daringer, N. M., Freije, C. A., Myhrvold, C., Bhattacharyya, R. P., Livny, J., Regev, A., Koonin, E. V., Hung, D. T., Sabeti, P. C., Collins, J. J., & Zhang, F. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science, 356(6336), 438–442. https://doi.org/10.1126/science.aam9321
Xie, S., Tao, D., Fu, Y., Xu, B., Tang, Y., Steinaa, L., Hemmink, J. D., Pan, W., Huang, X., Nie, X., Zhao, C., Ruan, J., Zhang, Y., Han, J., Fu, L., Ma, Y., Li, X., Liu, X., & Zhao, S. (2022). Rapid Visual Crispr Assay: A Naked-Eye Colorimetric Detection Method For Nucleic Acids Based On Crispr/Cas12a And A Convolutional Neural Network. ACS Synthetic Biology, 11(1), 383–396. https://doi.org/10.1021/acssynbio.1c00474
Li, S. Y., Cheng, Q. X., Wang, J. M., Li, X. Y., Zhang, Z. L., Gao, S., Cao, R. B., Zhao, G. P., & Wang, J. (2018). CRISPR-Cas12a-assisted nucleic acid detection. Cell discovery, 4, 20. https://doi.org/10.1038/s41421-018-0028-z
Liang, M., Li, Z., Wang, W., Liu, J., Liu, L., Zhu, G., Karthik, L., Wang, M., Wang, K. F., Wang, Z., Yu, J., Shuai, Y., Yu, J., Zhang, L., Yang, Z., Li, C., Zhang, Q., Shi, T., Zhou, L., … Zhang, L. X. (2019). A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nature Communications, 10(1), 3672. https://doi.org/10.1038/s41467-019-11648-1
Zhao, C., Zheng, X., Qu, W., Li, G., Li, X., Miao, Y. L., Han, X., Liu, X., Li, Z., Ma, Y., Shao, Q., Li, H., Sun, F., Xie, S., & Zhao, S. (2017). CRISPR-offinder: A CRISPR guide RNA design and off-target searching tool for user-defined protospacer adjacent motif. International Journal of Biological Sciences, 13(12), 1470–1478. https://doi.org/10.7150/ijbs.21312
Tao, D., Liu, J., Nie, X., Xu, B., Tran-Thi, T. N., Niu, L., Liu, X., Ruan, J., Lan, X., Peng, G., Sun, L., Ma, Y., Li, X., Li, C., Zhao, S., & Xie, S. (2020). Application of crispr-cas12a enhanced fluorescence assay coupled with nucleic acid amplification for the sensitive detection of African swine fever virus. ACS Synthetic Biology, 9(9), 2339–2350. https://doi.org/10.1021/acssynbio.0c00057
Qian, C., Wang, R., Wu, H., Zhang, F., Wu, J., & Wang, L. (2019). Uracil-mediated new photospacer-adjacent motif of Cas12a to realize visualized DNA detection at the single-copy level free from contamination. Analytical Chemistry, 91(17), 11362–11366. https://doi.org/10.1021/acs.analchem.9b02554
Swarts, D. C., & Jinek, M. (2019). Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Molecular Cell, 73(3), 589-600.e4. https://doi.org/10.1016/j.molcel.2018.11.021
Gao, X., Sun, B., & Guan, Y. (2019). Pullulan reduces the non-specific amplification of loop-mediated isothermal amplification (LAMP). Analytical and Bioanalytical Chemistry, 411(6), 1211–1218. https://doi.org/10.1007/s00216-018-1552-2
Quyen, T. L., Nordentoft, S., Vinayaka, A. C., Ngo, T. A., Engelsmenn, P., Sun, Y., Madsen, M., Bang, D. D., & Wolff, A. (2019). A sensitive, specific and simple loop mediated isothermal amplification method for rapid detection of campylobacter spp in broiler production. Frontiers in Microbiology, 10, 2443. https://doi.org/10.3389/fmicb.2019.02443
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., & Zhang, F. (2019). SHERLOCK: nucleic acid detection with CRISPR nucleases. Nature Protocols, 14(10), 2986–3012. https://doi.org/10.1038/s41596-019-0210-2
Ballester, M., Castelló, A., Ramayo-Caldas, Y., & Folch, J. M. (2013). A quantitative real-time PCR method using an X-linked gene for sex typing in pigs. Molecular Biotechnology, 54(2), 493–496. https://doi.org/10.1007/s12033-012-9589-5
Acknowledgements
This work was supported by the Natural Science Foundation of China (32072685), the Fund of Modern Industrial Technology System of Pig (CARS-35), and the Key Laboratory of Animal Embryo Engineering and Molecular Breeding of Hubei Province (KLAEMB-2020-05).
Author information
Authors and Affiliations
Contributions
SSX, DGT, and JJL: conceptualization. JJL and DGT: Methodology software. JJL and YJ: validation. QSL, PG, and JX: formal analysis. EMK: investigation. DGT, JJL and QSL: resources. SSX and LLN: data curation. JXR and YLM: writing—original draft preparation. SSX, JJL, DGT and QSL: writing—review and editing, SSX, DGT and EMK: visualization. DGT, BRX and QSL: supervision and project administration. SSX and LLN: funding acquisition. SSX All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tao, D., Liu, J., Li, Q. et al. A Simple, Affordable, and Rapid Visual CRISPR-Based Field Test for Sex Determination of Earlier Porcine Embryos and Pork Products. Mol Biotechnol 65, 263–272 (2023). https://doi.org/10.1007/s12033-022-00532-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00532-x