Abstract
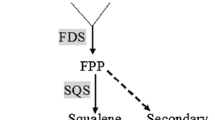
A farnesyl diphosphate synthase (FPS) cDNA and promoter region was cloned from Sanghuangporus baumii. The gene contains a 150-bp 5′-untranslated region (UTR), a 154-bp 3′-UTR, and a 1062-bp open reading frame (ORF) encoding a 354 amino acid polypeptide. The FPS-DNA includes three exons (nucleotides 1 –123, 184–321, and 505–1305) and two introns (nucleotides 124–183 and 322–504). The FPS protein has a molecular weight of 40.73 kDa, it is hydrophilic with a theoretical isoelectric point of 5.13, and the secondary and three-dimensional structure were analysed. There is a transcription start site at nucleotides 1318–1368 of the promoter, which includes typical eukaryotic promoter elements (TATA Box, CAAT Box, ARBE, AT-rich element, G-box, MBS, Sp1, LTR). FPS was expressed in Escherichia coli BL21, and the recombinant protein (63.41 kDa) was subjected to dodecyl sulphate, sodium salt-polyacrylamide gel electrophoresis (SDS-PAGE). FPS transcription was measured during different developmental stages, and expression in 11 and 13 days mycelia was upregulated 49.3-fold and 125.4-fold, respectively, compared with 9 days mycelia controls. Through analysing, S. baumii triterpenoid content was correlated with the transcription level of FPS during different development stages, and the triterpenoid content peaked at day 15 (7.21 mg/g).







Similar content being viewed by others
References
Zhou, L.-W., Vlasak, J., Decock, C., Assefa, A., Stenlid, J., Abate, D., et al. (2015). 第十二届海峡两岸真菌学学术研讨会大会手册壁报论文摘要集 (Global diversity and taxonomy of the Inonotus linteus complex (Hymenochaetales, Basidiomycota): Sanghuangporus gen.nov. Tropicoporus excentrodendri and T guanacastensis gen.et spp.nov. and 17 new combinations). Fungal Diversity 77(1), 335–347.
Sun, J., Chen, Q. J., Zhu, M. J., Wang, H. X., & Zhang, G. Q. (2014). An extracellular laccase with antiproliferative activity from the sanghuang mushroom Inonotus baumii. Journal of Molecular Catalysis B,99, 20–25.
Wang, F. F., Shi, C., Yang, Y., et al. (2018). Medicinal mushroomPhellinus igniarius induced cell apoptosis in gastric cancer SGC-7901 through a mitochondria-dependent pathway. Biomedicine & Pharmacotherapy,102, 18–25.
Ge, Q., Mao, J. W., Zhang, A. Q., Wang, Y. J., & Sun, P. L. (2013). Purification, chemical characterization, and antioxidant activity of a polysaccharide from the fruiting bodies of sanghuang mushroom (Phellinus baumii Pilát). Food Science and Biotechnology,22(2), 301–307.
Taddesse, Y., Lee, W. M., Ko, D., Park, S. C., Cho, J. Y., Park, H. J., et al. (2013). Phellinus baumii ethyl acetate extract alleviated collagen type II induced arthritis in DBA/1 mice. Journal of Natural Medicines,67(4), 807–813.
Wu, N. (2013). 桑黄子实体中抗衰老化合物的筛选及作用机制的初步探究(Screening the anti-aging active Components from the fruit of Phellinusbaumii and study on the mechanism of active compounds). Shanghai: Shanghai Normal University, PhD. Chinese.
Kim, D. I., Kim, K. S., Kang, J. H., & Kim, H. J. (2013). Effect of Phellinus baumii-biotransformed soybean powder on lipid metabolism in rats. Preventive Nutrition and Food Science,18(2), 98–103.
Xue, Q., Sun, J., Zhao, M. W., Zhang, K., & Lai, R. (2011). Immunostimulatory and anti-tumor activity of a water-soluble polysaccharide from Phellinus baumii mycelia. World Journal of Microbiology and Biotechnology,27(5), 1017–1023.
Wang, Y. Y., Ma, H., Ding, Z. C., Yang, Y., Wang, W. H., Zhang, H. N., et al. (2019). baumii. International Journal of Biological Macromolecules,123, 201–209.
Taji, S., Yamada, T., Wada, S., Tokuda, H., Sakuma, K., & Tanaka, R. (2008). Lanostane-type triterpenoids from the sclerotia of Inonotus obliquus possessing anti-tumor promoting activity. European Journal of Medicinal Chemistry,43(11), 2373–2379.
Zhang, L. F., Sun, T. T., & Zou, L. (2015). 鲍姆纤孔菌总三萜的提取及其体外抗乳腺癌细胞 (MCF-7)活性 (Extraction of total triterpenoids from Inonotus baumii and its inhibitory activity on breast cancer cells (MCF-7) in vitro). Drug Evaluation Research,38(5), 497–502.
Zhang, G. L., Si, J., Tian, X. M., & Wang, J.-P. (2017). 真菌激发子对桑黄胞内代谢产物积累的影响 (The effects of fungal elicitor on the accumulation of Sanghuangporus sanghuang intracellular metabolites). Mycosystema,36(4), 482–491.
Sun, T. T., Zou, L., Zhang, L. F., Zhang, J., & Wang, X. (2017). Methyl jasmonate induces triterpenoid biosynthesis in Inonotus baumii. Biotechnology & Biotechnological Equipment,31(2), 312–317.
Hemmerlin, A., Rivera, S. B., Erickson, H. K., & Poulter, C. D. (2003) Enzymes encoded by the farnesyl diphosphate synthase genefamily in the big sagebrush Artemisia tridentata ssp. spiciformis. Journal of Biological Chemistry, 278(34), 32132–32140.
Rodríguez-Concepción, M., & Boronat A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant physiology, 130(3), 1079–1089.
Hunter, W. N. (2007). The non-mevalonate pathway of isoprenoid precursor biosynthesis. The Journal of Biological Chemistry,282(30), 21573–21577.
Liao, Z. H., Chen, M., Gong, Y. F., Li, Z. G., Zuo, K. J., Wang, P., et al. (2006). A new farnesyl diphosphate synthase gene from Taxus media rehder: Cloning, characterization and functional. Journal of Integrative Plant Biology,48(6), 692–699.
Zjawiony, J. K. (2004). Biologically active compounds from Aphyllophorales (polypore) fungi. Journal of Natural Products,67(2), 300–310.
Lange, B. M., Rujan, T., Martin, W., & Croteau, R. (2000). Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proceedings of the National Academy of Sciences of the United States of America,97(24), 13172–13177.
Poulter, C. D. (2006). Farnesyl diphosphate synthase. A paradigm for understanding structure and function relationships in E-polyprenyl diphosphate synthases. Phytochemistry Reviews, 5 (1), 17–26.
Chen, D. H., Ye, H. C., & Li, G. F. (2000). Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Science,155, 179–185.
Liu, M. J., Yu, Y. L., Jiang, S., et al. (2018). 珠子参中法尼基焦磷酸合酶(FPS)对皂苷生物合成的影响研究 (Effect of Farnesyl-pyrophosphate Synthase( FPS) on the Biosynthesis of Saponins in Panax japonicas). Bulletin of Botanical Research,38(4), 611–618.
Fei, Y., Li, N., Zhang, D. H., & Xu, J. W. (2019). Increased production of ganoderic acids by overexpression of homologous farnesyl diphosphate synthase and kinetic modeling of ganoderic acid production in Ganoderma lucidum. Microbial Cell Factories,18, 115.
Park, H. W., Kim, O. T., Hyun, D. Y., Kim, T. B., Kim, J. U., Kim, Y., et al. (2013). Overexpression of farnesyl diphosphate synthase by introducing cafps gene in Panax ginseng C. A. mey. Korean Journal of Medicinal Crop Science, 21(1), 32–38.
Gupta, P., Akhtar, N., Tewari, S. K., Sangwan, R. S., & Trivedi, P. K. (2011). Differential expression of farnesyl diphosphate synthase gene from with ania somniferain different chemotypes and in response to elicitors. Plant Growth Regulation,65(1), 93–100.
Ding, Y. X., -Yang, X., Shang, C. H., Ren, A., Shi, L., Li, Y. X., et al. (2008). Molecular cloning, characterization, and differential expression of a farnesyl-diphosphate synthase gene from the basidiomycetous fungus Ganoderma lucidum . Journal of the Agricultural,72(6), 9.
Qi, Y., Liu, C., Sun, X., Qiu, L., & Shen J. (2017). The identification of transcriptional regulation related gene of laccase poxc through yeast one-hybrid screeningfrom Pleurotus ostreatus. Fungal Biology 2017, 121(11), 905–910.
Zhang, W. B., Wang, J. G., Li, Z. K., Yang, L. Q., Qin, J., Xiang, Z. H., et al. (2014). 药用真菌桑黄的研究进展 (Progress of studies on medicinal fungus Phellinus). China Journal of Chinese Materia Medica,39(15), 2838–2845.
Jia, L. (2016). 茯苓总三萜免疫抑制及诱导人结肠癌RKO细胞凋亡的硏究 (Study of immunosuppressive activity and apoptosis-inducing effect of total triterpenoids from Poria cocos on human colorectal carcinoma). Guangdong: Southern Medical University, PhD. Chinese.
Guruprasad, K., Reddy, B. V., & Pandit, M. W. (1990). Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Engineering, Design and Selection,4(2), 155–161.
Garnier, B. J., Gibrat, J. F., & Robson, B. (1996). GOR secondary structure prediction method version IV. Methods in Enzymology,266, 540–553.
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research,46(1), 296–303.
Guex, N., Peitsch, M. C., & Schwede, T. (2010). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis,30(S1), S162–S173.
Bienert, S., Waterhouse, A., & de Beer, T. A. P. (2017). The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Research,45(1), D313–D319.
Benkert, P., Biasini, M., & Schwede, T. (2011). Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics,27(3), 343–350.
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., & Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Scientific Reports,7(1), 10480.
Tachibana, A ., Yano, Y., Otani, S., Nomura, N., Sako Y., & Taniguchi, M. (2000). Novel prenyltransferase gene encoding farnesylgeranyl diphosphate synthase from a hyperthermophilic archaeon, Aeropyrum pernix. Molecularevolution with alteration in product specificity. European Journal of Biochemistry, 267(2), 321.
Anderson, M. S., Yarger, J. G., Burck, C. L., & Poulter C. D. (1989). Farnesyl diphosphate synthetase. Molecular cloning, sequence, and expression of an essential gene from Saccharomyces cerevisiae. Journal of Biological Chemistry, 264(32), 19176–19184.
Hemmerlin, A., Rivera, S. B., Erickson, H. K., & Poulter C. D. (2003). Enzymes encoded by the farnesyl diphosphate synthase gene family in the big Sagebrush Artemisia tridentata ssp. spiciformis. Journal of Biological Chemistry, 278(34), 32132–32140.
Cervantes-Cervantes, M. (2006). Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant Physiology,141(1), 220–231.
Ding, Y. X. (2013). 灵芝法尼基焦磷酸合酶基因的克隆和表达特性研究 (Molecular cloning, characterization, and differential expression of a farnesyl-diphosphate synthase gene from the basidiomycetous fungus Ganoderma lucidum). Nanjing: Nanjing Agricultural University. PhD Dissertations. Chinese.
Zhu, D. Q. (2016). 基因表达过程中涨落与延迟作用机制的研究 (The study of fluctuation and delay effects in gene expression process), Anhui: University of Science and Technology of China. Chinese: PhD.
Zhang, C. B., Sun, H. X., & Gong, Z. J. (2000). 植物萜类化合物的天然合成途径及其相关合酶 (Plant terpenoid natural metabolism pathways and their synthases). Plant Physiology Journal,43(04), 779–786.
Acknowledgements
This research was supported by the Fundamental Research Funds for the Central Universities, China (2572017CF01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, X., Sun, T., Sun, J. et al. Molecular Cloning, Characterisation, and Heterologous Expression of Farnesyl Diphosphate Synthase from Sanghuangporus baumii. Mol Biotechnol 62, 132–141 (2020). https://doi.org/10.1007/s12033-019-00231-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-019-00231-0