Abstract
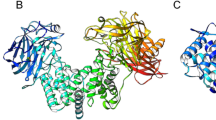
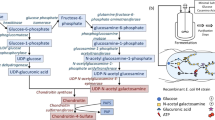
Glycosaminoglycans (GAGs) and their low-molecular weight derivates have received considerable interest in terms of their potential clinical applications, and display a wide variety of pharmacological and pharmacokinetic properties. Structurally distinct GAG chains can be prepared by enzymatic depolymerization. A variety of bacterial chondroitin sulfate (CS) lyases have been identified, and have been widely used as catalysts in this process. Here, we identified a putative chondroitin AC exolyase gene, AschnAC, from an Arthrobacter sp. strain found in a CS manufacturing workshop. We expressed the enzyme, AsChnAC, recombinantly in Escherichia coli, then purified and characterized it in vitro. The enzyme indeed displayed exolytic cleavage activity toward HA and various CSs. Removing the putative N-terminal secretion signal peptide of AsChnAC improved its expression level in E. coli while maintaining chondroitin AC exolyase activity. This novel catalyst exhibited its optimal activity in the absence of added metal ions. AsChnAC has potential applications in preparation of low-molecular weight GAGs, making it an attractive catalyst for further investigation.





Similar content being viewed by others
References
Varki, A., Cummings, R. D., Esko, J. D., Freeze, H. H., Stanley, P., Bertozzi, C. R., et al. (Eds.). (2009). Essentials of glycobiology (2nd ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
DeAngelis, P. L., Liu, J., & Linhardt, R. J. (2013). Chemoenzymatic synthesis of glycosaminoglycans: Re-creating, re-modeling and re-designing nature’s longest or most complex carbohydrate chains. Glycobiology, 23(7), 764–777.
Gandhi, N. S., & Mancera, R. L. (2008). The structure of glycosaminoglycans and their interactions with proteins. Chemical Biology & Drug Design, 72(6), 455–482. https://doi.org/10.1111/j.1747-0285.2008.00741.x.
Taylor, K. R., & Gallo, R. L. (2006). Glycosaminoglycans and their proteoglycans: Host-associated molecular patterns for initiation and modulation of inflammation. The FASEB Journal, 20(1), 9–22. https://doi.org/10.1096/fj.05-4682rev.
Xu, D., & Esko, J. D. (2014). Demystifying heparan sulfate-protein interactions. Annual Review of Biochemistry, 83(1), 129–157. https://doi.org/10.1146/annurev-biochem-060713-035314.
Pantazaka, E., & Papadimitriou, E. (2014). Chondroitin sulfate-cell membrane effectors as regulators of growth factor-mediated vascular and cancer cell migration. Biochimica et Biophysica Acta (BBA)-General Subjects, 1840(8), 2643–2650. https://doi.org/10.1016/j.bbagen.2014.01.009.
Li, P., Sheng, J., Liu, Y., Li, J., Liu, J., & Wang, F. (2013). Heparosan-derived heparan sulfate/heparin-like compounds: One kind of potential therapeutic agents. Medicinal Research Reviews, 33(3), 665–692. https://doi.org/10.1002/med.21263.
Li, L., Li, Y., Ijaz, M., Shahbaz, M., Lian, Q., & Wang, F. (2015). Review on complement analysis method and the roles of glycosaminoglycans in the complement system. Carbohydrate Polymers, 134, 590–597. https://doi.org/10.1016/j.carbpol.2015.08.028.
Migliore, A., & Procopio, S. (2015). Effectiveness and utility of hyaluronic acid in osteoarthritis. Clinical Cases in Mineral and Bone Metabolism, 12(1), 31–33. https://doi.org/10.11138/ccmbm/2015.12.1.031.
Liu, J., & Pedersen, L. C. (2006). Anticoagulant heparan sulfate: Structural specificity and biosynthesis. Applied Microbiology and Biotechnology, 74(2), 263–272. https://doi.org/10.1007/s00253-006-0722-x.
Zhang, Q., Li, J., Liu, C., Song, C., Li, P., Yin, F., et al. (2015). Protective effects of low molecular weight chondroitin sulfate on amyloid beta (Aβ)-induced damage in vitro and in vivo. Neuroscience, 305, 169–182. https://doi.org/10.1016/j.neuroscience.2015.08.002.
Bisio, A., Vecchietti, D., Citterio, L., Guerrini, M., Raman, R., Bertini, S., et al. (2009). Structural features of low-molecular-weight heparins affecting their affinity to antithrombin. Thrombosis and Haemostasis, 102(5), 865–873. https://doi.org/10.1160/TH09-02-0081.
Merli, G. J., & Groce, J. B. (2010). Pharmacological and clinical differences between low-molecular-weight heparins. Pharmacy and Therapeutics, 35(2), 95–105.
Pempe, E. H., Xu, Y., Gopalakrishnan, S., Liu, J., & Harris, E. N. (2012). Probing structural selectivity of synthetic heparin binding to stabilin protein receptors. The Journal of Biological Chemistry, 287(25), 20774–20783. https://doi.org/10.1074/jbc.M111.320069.
Ernst, S., Langer, R., Cooney, C. L., & Sasisekharan, R. (1995). Enzymatic degradation of glycosaminogIycans. Critical Reviews in Biochemistry and Molecular Biology, 30(5), 387–444. https://doi.org/10.3109/10409239509083490.
Lunin, V. V., Li, Y., Linhardt, R. J., Miyazono, H., Kyogashima, M., Kaneko, T., et al. (2004). High-resolution crystal structure of arthrobacter aurescens chondroitin AC Lyase: An enzyme-substrate complex defines the catalytic mechanism. Journal of Molecular Biology, 337(2), 367–386. https://doi.org/10.1016/j.jmb.2003.12.071.
Lombard, V., Bernard, T., Rancurel, C., Brumer, H., Coutinho, P. M., & Henrissat, B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. The Biochemical Journal, 432(3), 437–444. https://doi.org/10.1042/BJ20101185.
Garron, M.-L., & Cygler, M. (2010). Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology, 20(12), 1547–1573. https://doi.org/10.1093/glycob/cwq122.
Linhardt, R. J., Avci, F. Y., Toida, T., Kim, Y. S., & Cygler, M. (2006). CS lyases: Structure, activity, and applications in analysis and the treatment of diseases. Advances in Pharmacology, 53, 187–215.
Hernáiz, M. J., & Linhardt, R. J. (2001). Degradation of chondroitin sulfate and dermatan sulfate with chondroitin lyases. Methods in Molecular Biology, 171, 363–371. https://doi.org/10.1385/1-59259-209-0:363.
Huang, W., Boju, L., Tkalec, L., Su, H., Yang, H. O., Gunay, N. S., et al. (2001). Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis. Biochemistry, 40(8), 2359–2372.
Capila, I., Wu, Y., Rethwisch, D. W., Matte, A., Cygler, M., & Linhardt, R. J. (2002). Role of arginine 292 in the catalytic activity of chondroitin AC lyase from Flavobacterium heparinum. Biochimica et Biophysica Acta, 1597(2), 260–270.
Kale, V., Friðjónsson, Ó., Jónsson, J. Ó., Kristinsson, H. G., Ómarsdóttir, S., & Hreggviðsson, G. Ó. (2015). Chondroitin lyase from a marine Arthrobacter sp. MAT3885 for the production of chondroitin sulfate disaccharides. Marine Biotechnology, 17(4), 479–492. https://doi.org/10.1007/s10126-015-9629-9.
Fang, Y., Yang, S., Fu, X., Xie, W., Li, L., Liu, Z., et al. (2019). Expression, purification and characterization of chondroitinase AC II from marine bacterium Arthrobacter sp. CS01. Marine Drugs, 17(3), 185. https://doi.org/10.3390/md17030185.
Ke, T., Zhangfu, L., Qing, G., Yong, T., Hong, J., Hongyan, R., et al. (2005). Isolation of Serratia marcescens as a chondroitinase-producing bacterium and purification of a novel chondroitinase AC. Biotechnology Letters, 27(7), 489–493. https://doi.org/10.1007/s10529-005-2538-7.
Shim, K.-W., & Kim, D.-H. (2008). Cloning and expression of chondroitinase AC from bacteroides stercoris HJ-15. Protein Expression and Purification, 58(2), 222–228. https://doi.org/10.1016/j.pep.2007.11.014.
Yin, F.-X., Wang, F.-S., & Sheng, J.-Z. (2016). Uncovering the catalytic direction of chondroitin AC exolyase from the reducing end towards the non-reducing end. Journal of Biological Chemistry, 291(9), 4399–4406. https://doi.org/10.1074/jbc.C115.708396.
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., & Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6(5), 343–345. https://doi.org/10.1038/nmeth.1318.
Wang, T.-T., Zhu, C.-Y., Zheng, S., Meng, C.-C., Wang, T.-T., Meng, D.-H., et al. (2018). Identification and characterization of a chondroitin synthase from Avibacterium paragallinarum. Applied Microbiology and Biotechnology, 102(11), 4785–4797. https://doi.org/10.1007/s00253-018-8926-4.
Robert, X., & Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42(W1), W320–W324. https://doi.org/10.1093/nar/gku316.
Bordoli, L., Kiefer, F., Arnold, K., Benkert, P., Battey, J., & Schwede, T. (2009). Protein structure homology modeling using SWISS-MODEL workspace. Nature Protocols, 4(1), 1–13. https://doi.org/10.1038/nprot.2008.197.
Maiti, R., Van Domselaar, G. H., Zhang, H., & Wishart, D. S. (2004). SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Research, 32, W590–W594. https://doi.org/10.1093/nar/gkh477.
Ochman, H., Gerber, A. S., & Hartl, D. L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics, 120(3), 621–623.
Park, K.-B., & Oh, S.-H. (2007). Cloning, sequencing and expression of a novel glutamate decarboxylase gene from a newly isolated lactic acid bacterium, Lactobacillus brevis OPK-3. Bioresource Technology, 98(2), 312–319. https://doi.org/10.1016/j.biortech.2006.01.004.
Liu, Q., Liu, H.-C., Zhou, Y.-G., & Xin, Y.-H. (2019). Genetic diversity of glacier-inhabiting Cryobacterium bacteria in China and description of Cryobacterium zongtaii sp. nov. and Arthrobacter glacialis sp. nov. Systematic and Applied Microbiology, 42(2), 168–177. https://doi.org/10.1016/j.syapm.2018.10.005.
Castrignanò, T., DE Meo, P. D., Cozzetto, D., Talamo, I. G., & Tramontano, A. (2006). The PMDB protein model database. Nucleic Acids Research, 34(suppl_1), D306–D309. https://doi.org/10.1093/nar/gkj105.
Guo, X., Shi, Y., Sheng, J., & Wang, F. (2014). A novel hyaluronidase produced by Bacillus sp. A50. PLoS ONE, 9(4), e94156. https://doi.org/10.1371/journal.pone.0094156.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673–4680. https://doi.org/10.1093/nar/22.22.4673.
Funding
Funding was provided by National Natural Science Foundation of China (Grant No. 31770845)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, LZ., Shi, CQ., Yin, FX. et al. Cloning and Characterization of a Chondroitin AC Exolyase from Arthrobacter sp. SD-04. Mol Biotechnol 61, 791–800 (2019). https://doi.org/10.1007/s12033-019-00208-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-019-00208-z