Abstract
We have developed a transencapsidated vaccine delivery system based on the insect virus, Flock House virus (FHV). FHV is attractive due to its small genome size, simple organization, and nonpathogenic characteristics. With the insertion of a Tobacco mosaic virus (TMV) origin of assembly (Oa), the independently replicating FHV RNA1 can be transencapsidated by TMV coat protein. In this study, we demonstrated that the Oa-adapted FHV RNA1 transencapsidation process can take place in planta, by using a bipartite plant expression vector system, where TMV coat protein is expressed by another plant virus vector, Foxtail mosaic virus (FoMV). Dual infection in the same cell by both FHV and FoMV was observed. Though an apparent classical coat protein-mediated resistance repressed FHV expression, this was overcome by delaying inoculation of the TMV coat protein vector by 3 days after FHV vector inoculation. Expression of the transgene marker in animals by these in vivo-generated transencapsidated nanoparticles was confirmed by mouse vaccination, which also showed an improved vaccine response compared to similar in vitro-produced vaccines.
Similar content being viewed by others
Introduction
Virus-based nanoparticles have been extensively explored as a vaccine delivery strategy due to their typically higher immunogenicity compared with unassembled vaccine antigens [1, 2], their potential to serve as their own adjuvant [1–3], and their greater safety and potentially relatively lower cost of protection compared to traditional vaccines [4]. Virus-like particles (VLPs) display vaccine antigen on their surface and can be produced by the self-assembly of viral coat protein subunits expressed in a heterologous host, such as bacteria [5] or plants [6], or in mammalian cells [7]. An alternative to VLPs is to use viral coat protein to encapsidate the RNA of another virus, with the RNA expressing the vaccine antigen once delivered to the target cell. In this way, the viral RNA can be packaged in an especially resistant nanoparticle similar to a VLP. The potential advantage of this strategy over VLPs is the activation of innate immunity by viral replication [8–10].
Among numerous trials using viral nanoparticles for antigen delivery, Tobacco mosaic virus (TMV) nanoparticles seem to hold special promise. TMV virions are characterized by great stability and low-cost production [11], and a recent study suggests that the human population has already been extensively exposed to TMV coat antigen through exposure to food and tobacco sources [12]. Furthermore, extensive data show that pre-existing immunity to TMV coat does not disrupt boosting of either cytotoxic T lymphocyte (CTL), [13, 14] or antibody target antigens [15, 16]. Lastly, TMV virions are extremely stable, remaining infective for over a century at room temperature [17]. TMV exhibits robust expression in plants at up to 5–10 % dry weight and is easy to purify at the commercial scale [11].
Consequently, TMV nanoparticles have been explored as a VLP epitope platform. The highly uniform repeated organization of 2,130 copies of coat protein subunits and the associated strong cross-linking pattern provide greatly improved efficacy to deliver antigens to antigen-presenting cells. Various studies have validated that TMV-antigen conjugation can induce B cell activation and raise antibody titers [15, 18, 19], even when the conjugates are poorly immunogenic, such as carbohydrates [20]. Furthermore, TMV uptake by dendritic cells is rapid and efficient [14, 18], and peptide-presenting TMV nanoparticles were proven to be able to elicit T cell responses with augmented interferon gamma (IFNγ) levels [14]. We have also previously successfully tested ovalbumin-conjugated TMV vaccines, as well as a bivalent TMV vaccine displaying both mouse melanoma-associated CTL epitopes p15e and tyrosinase-related protein 2 (Trp2) peptides [13]. Immunization resulted in a significantly improved survival after lethal tumor challenge. A recent study also demonstrated TMV’s great potential to be used in stand-alone or prime-boost dendritic cell activation strategies [18].
In addition to utilizing TMV as a VLP to present surface epitopes, development has also proceeded with TMV coat protein-encapsidated RNA vaccines. In previous experiments, we have produced and tested Semliki Forest virus (SFV) RNA encapsidated with TMV coat protein in vitro. Attenuated SFV was modified by insertion of a TMV origin of assembly to produce, in vitro, rod-shaped virus particles that resembled TMV [21] by mixing SFV-Oa RNA with purified TMV coat protein. Vaccination with SFV-Oa encoding the model antigen beta-galactosidase (bGal) resulted in boosted antibody responses to bGal protein, demonstrating that TMV-encapsidated RNA was translated and was antigenic in the absence of adjuvant and, further, that the presence of the TMV Oa did not disrupt SFV replication functions. However, as a common phenomenon of pathogenic RNA virus vaccines [22], SFV-Oa RNA induced apoptosis in infected cells, which may limit duration of antigen exposure and reduce immune activation to transgene-encoded antigens.
To improve on our previous results with SFV, we applied TMV encapsidation to the RNA of the nonpathogenic insect virus, Flock House virus (FHV), which is capable of replicating in human cells. The advantages of FHV include a bipartite genome, where the polymerase is encoded by the independently replicating RNA 1 and the structural capsid gene is encoded by RNA 2, allowing for easy manipulation of the RNA1 genome for vaccine development and the separation of replication from packaging. We have already tested in vitro-assembled TMV–FHV particles and have shown that TMV Oa did not disrupt FHV viral replication, using an enhanced green fluorescent protein (eGFP) transgene to monitor replication and expression in mammalian cells [23]. However, the limitations of in vitro encapsidation remain with this system, namely the cost of RNA synthesis and potentially reduced translation due to inefficient in vitro 5′ capping.
To overcome these limitations, we explored an in planta strategy for producing viral RNA in vivo. For the present study, we hypothesized that FHV RNA, which replicates well in mammalian [24] and plant cells [25] but is not a pathogen of either, could be encapsidated in planta if sufficient TMV coat proteins were provided in trans. We further predicted that in planta-produced nanoparticles would be able to express transgene after animal vaccination and will be comparatively more immunogenic than in vitro nanoparticles due to natural 5′ capping. As described in the following report, we used a 35S promoter to express FHV-eGFP-Oa RNA and the plant viral vector Foxtail mosaic virus (FoMV) to express TMV coat protein in quantities sufficient for encapsidation of FHV RNA in agroinoculated Nicotiana benthamiana plants. Replication of functional FHV-eGFP-Oa was observed as an unusually strong eGFP fluorescence, and near wild-type levels of TMV coat protein were produced by co-delivered FoMV vector. We observed virion particles of the typical TMV morphology as a final product. When these nanoparticles were used to vaccinate mice, the expression of eGFP transgene was confirmed by an anti-eGFP immune response greater than that observed for in vitro encapsidated control particles. This is the first report of in planta transencapsidated nanoparticles and represents the first step toward producing a commercially viable vaccine of this type.
Materials and Methods
Construction of T7/FHV-C2-GFP Vector and Expression in Mammalian Cells
The plasmid containing the FHV RNA1 expression cassette was kindly provided by Dr. A. Ball. It is a T7 promoter-driven plasmid containing the RNA1 portion of the FHV genome and was previously described [26]. A polylinker, CTCGAGGCGATCGCCTGCAG, encompassing the three restriction sites XhoI, AsiSI, and PstI, was cloned into one of four insertion sites: C1, nt. 3034; C2, nt. 3037; C3, nt. 2731; and C4, nt. 3055, and confirmed by direct sequencing. Enhanced green fluorescent protein (eGFP) ORF was then cloned into these sites via XhoI and PstI to create T7/FHV-C- [1–4] -GFP constructs (Fig. 1a). To confirm stability of the eGFP-modified FHV viral RNA, full-length RNA transcripts were generated from the T7/FHV-C-GFP DNA in vitro via a T7 promoter kit (mMessage mMachine, Ambion, TX). 2 µg RNA was used to transfect BHK-21 cells with DMRIE-C (Invitrogen, Carlsbad, CA). Transfected cells were incubated at 37 °C for 4 h, after which fresh growth media were used to replace transfection media. Cells were then placed at 28 °C for 24 h. Expression of fluorescence was confirmed using a Nikon Eclipse TS100 microscope and NIS-elements imaging software. Cells were observed for 2 days post-transfection.
FHV viral vector constructs for expression in mammalian cells. a Two constructs, C2 and C4, differing in the insertion site for eGFP. B2, FHV silencing suppressor; Rbz, HDV ribozyme for precise viral RNA 3′ end excision. b Expression of T7/FHV-C2-GFP (left) and T7/FHV-C4-GFP (right) in BHK21 cells (Color figure online)
In Planta Expression Vectors
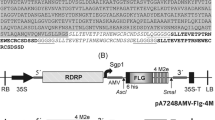
In order to express FHV in plants, full-length FHV viral vector sequence was transferred from T7/FHV-C2-GFP (Fig. 1a) and placed between the StuI/XbaI sites of the plant binary vector JL22 [27] to create 35S/FHV-C2 (Fig. 2a). To allow Oa insertion, additional restriction sites were introduced on either side of the eGFP ORF by amplifying the eGFP ORF with an upstream primer containing XhoI/AscI and a downstream primer containing AvrII/PstI and then reinserting this product into 35S/FHV-C2 between the XhoI and PstI sites. TMV Oa (95 bp: TMV nts. 5432–5527) [28] was inserted upstream or downstream of the eGFP ORF to create 35S/FHVC2-o1 and -o2, respectively (Fig. 2a). PCR with a primer containing a mutated eGFP ORF stop codon was used to create 35S/FHVC2-o3.
FHV viral vector constructs for expression in plants. The C2 and C4 constructs from Fig. 1 were provided with the 35S plant expression promoter and the TMV origin of assembly (Oa) to allow for encapsidation. a In the C2 series, TMV Oa was added at different positions in C2-o1, and -o2, and in -o3 the eGFP native C-terminal stop codon was preserved. B2′ stands for the B2 ORF C-terminal remaining after eGFP insertion. b The C4-2sg has a duplicated subgenomic promoter to express unfused versions of eGFP and B2 silencing suppressor. The eGFP ORF is between the duplicated subgenomic promoters in C4-2sg, but follows the final subgenomic promoter in 2sg2 construct. The 2sg2 constructs retain the B2 C-terminus (B2″) following the eGFP ORF to mimic the 3′ end of the native FHV. c The FECT/TMV CP construct and the JL6/p19 were used as co-agroinoculants and provided coat protein and silencing suppressor, respectively
Several modifications were made to improve eGFP expression. T7/FHV-C4-2sg was created to maintain B2 expression, by duplicating the 3′ end of FHV RNA1 (nt. 2518–3055) and inserting it after the eGFP open reading frame in T7/FHV-C4-GFP. 35S/FHV-C4-2sg (Fig. 2b) was generated by transferring the viral sequence into JL22 [27], as outlined above. To express both FHV B2 and eGFP separately, a 498 bp DNA segment was synthesized (gBlock, IDT, Coralville, IA) and inserted between the XhoI and PstI sites in 35S/FHV-C4. This segment contained a stop codon in the B2 ORF, 10 bp of the FHV 3′ UTR for any potential required context for B2 ORF expression, the 95 bp TMV Oa, a repeat of the presumed B2 subgenomic promoter (FHV 2480–2809, including 69 bp past the B2 start) to drive eGFP expression, and a start codon and insertion sites for eGFP. To recreate a more FHV authentic 3′ region following the eGFP ORF, the final 24 bases of B2 ORF was added downstream of the eGFP stop codon, to yield the final construct: 35S/FHV2sg2 (Fig. 2b). All recombinant DNA methods and suppliers for the plant constructs were as previously described [29].
Agroinoculation and Visualization
Nicotiana benthamiana plants were grown and agroinoculated as previously described [29]. Excised eGFP-fluorescent leaves were visualized using a blue light Dark Reader (Clare Chemical, Dolores, CO, USA). The defective interfering construct DI638/wtGFP [30] was a gift from A. Rao (UC Riverside), and those inoculations were visualized with a hand-held UVL-56 lamp (UVProducts, Upland, CA, USA).
Relative fluorescence resulted by different FHV constructs was measured by grinding inoculated leaf tissue in 1× Phosphate Buffered Saline (PBS). The collected supernatant was assessed on a microplate reader (Thermo Fluoroskan Ascent FL), with black 96-well plate (COSTAR 3925, Corning Inc. NY). Filter set of 485 nm (excitation) and 538 nm (emission) was used in order to detect eGFP fluorescence.
Plant Protoplasts
Protoplasts were prepared from N. benthamiana leaves 4 days post-inoculation. Leaves were sliced into 2-mm strips and vacuum infiltrated with MMC buffer (13 % mannitol, 5 mM MES, 10 mM CaCl2, pH 5.8) containing 1 % Onozuka cellulase RS and 0.5 % Macerase (both from Phytotechnology Labs, Shawnee Mission, KS, USA) and gently rocked overnight. Protoplasts were mounted in MMC on a glass slide. Images were obtained as previously described [29].
Plant-Produced Nanoparticles
To purify nanoparticles, agroinoculated N. benthamiana leaves, 4–7 days p.i., were ground in a mortar in extraction buffer (50 mM sodium acetate, 0.86 M NaCl (5 % w/v), 0.04 % sodium metabisulfite, pH 5.0). Crude homogenate was filtered through cheesecloth and 8 % (v/v) n-butanol was added, and then incubated at room temperature for 15 min, and then centrifuged at 10,000×g for 15 min. The supernatant was decanted through cheesecloth, and nanoparticles were precipitated with PEG 8000 (EMD Millipore, USA) at 4 % on ice for 1 h, followed by centrifugation at 10,000×g for 10 min. The pellet was resuspended in a minimum of 10 mM phosphate buffer (pH 7.2) and then centrifuged at 16,000×g for 10 min. The supernatant was collected and nanoparticles were purified with an additional round of PEG precipitation. The final nanoparticle pellet was suspended in 10 mM phosphate buffer (pH 7.2) and stored at −20 °C. Protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce Biotechnology, Rockford, IL, USA).
Transmission electron microscopy was used to visualize purified nanoparticles on a JEOL JSM 1010 microscope. A 3 µl drop of nanoparticles was adsorbed onto 300-mesh formvar-coated grids (Electron Microscopy Sciences, PA, USA) for 1 min, drawn off, and stained with 1 % phosphotungstic acid (pH 7). Images were taken by XR16 TEM camera (Advanced Microscopy Techniques, MA, USA), and with AMT Image Capture Engine V602 (Advanced Microscopy Techniques, MA, USA), at 30,000× to 40,000× magnification.
In Vitro Nanoparticle Assembly and Vaccine Preparation
SFV-eGFP or FHV-eGFP RNA was transcribed from T7 plasmids using a capped RNA synthesis kit (mMessage mMachine; Ambion), quantitated by absorbance, and checked for integrity by gel electrophoresis. 50 µg of RNA was then incubated with 1.4 mg of TMV coat protein, prepared by a modified protocol as previously described [21]. Briefly, encapsidations were carried out using overnight incubation in a 0.05 M phosphate buffer (pH 7) at room temperature. Particles were recovered by PEG precipitation and quantitated by BCA assay (BioRad, Hercules, CA).
Vaccination and Immune Response Evaluation in Mice
BALB/c mice (Charles River, Hollister, CA) were housed at Touro University according to guidelines established in the Care and Use of Animals, and performed according to IACUC-approved protocols. Typically, three mice were given a 100–200 µl subcutaneous (s.c.) injection of 15 or 30 μg encapsidated product, or 15 µg eGFP protein as a positive control (Vector Labs), or PBS as a negative control. Vaccines were typically administered at 2-week intervals, and tail vein bleeds were taken at 10 days after vaccines 2 and 3 for enzyme-linked immunosorbent assay (ELISA) analysis.
The IgG immune response was determined by ELISA. 96-well microtiter plates (MaxiSorp; Nalge Nunc) were coated with 5 µg/ml eGFP protein (Vector Labs) in 50 mM carbonate/bicarbonate buffer (pH 9.6). After blocking with 2 % bovine serum albumin (BSA) in PBS, serial dilutions of the sera were added for 1 h, and the plates were washed and incubated for an additional hour with anti-mouse IgG Horse Radish Peroxidase (HRP)-conjugated secondary antibody (Southern Biotech) in PBS + BSA. Plates were developed using a tetramethyl benzidine substrate solution (TMB; BioFx), and the reactions were stopped by the addition of 1 N sulfuric acid. Plate absorbance was read at 450 nm in a 96-well plate spectrophotometer (Molecular Devices). Relative anti-eGFP titers reported were determined from a standard curve generated by a threefold serial dilution of a 100 ng/ml rabbit anti-eGFP polyclonal antibody (Sigma) detected with an anti-Rabbit-HRP secondary. Statistical analysis was carried out using Prism software (GraphPad), using unpaired t test with Welch’s correction.
Results
FHV Vector Expression in Mammalian Cells
FHV vectors were designed and tested for the expression of eGFP in BHK-21 cells. A cassette containing three restriction sites (XhoI, AsiI and PstI) was placed at the FHV C2 site [31], namely, immediately downstream of the polymerase/B1 stop codon, which is also six codons upstream from, and in phase with, the B2 stop codon (Fig. 1a). This FHV-C2 construct thus expresses a B2-eGFP-B2 fusion, with 99 amino acids of B2 upstream of eGFP and 6 amino acids of B2 at the C-terminus. Insertion at a second eGFP ORF insertion site, the C4 site, would produce the full B2 protein fused to the eGFP (Fig. 1a). eGFP expression was observed in mammalian cells (Fig. 1b) within 24 h post-transfection. Expression with both constructs peaked at 48 h and was maintained until 72 h, with approximately 15–20 % transfection efficiency. Fluorescence began to decrease after 72 h and gradually diminished over time. The C4 insertion construct gave reduced fluorescence compared to C2 (Fig. 1b).
Strong FHV/eGFP Expression in N. benthamiana After p19 Co-agroinoculation
The 35S/FHV-C2-GFP and 35S/FHV-C4-2sg constructs were made by transferring viral sequences from the mammalian vectors into plant binary vector pJL22 [27] between a cauliflower mosaic virus 35S promoter and 35S terminator (Fig. 2). Leaves agroinoculated with these constructs gave a weak fluorescence (Fig. 3), as did leaves inoculated with the positive FHV/wtGFP control, F1DI, comprising FHV RNA1 and DI638, the defective interfering RNA of FHV RNA2, carrying wtGFP [30]. However, when the silencing suppressor, p19 [32], was provided by co-agroinoculation, a much stronger fluorescence was observed (Fig. 3) which was much stronger than the F1DI + p19 control. Subsequently, p19 was included in all inoculations.
eGFP expression from constructs from Fig. 2. Agroinoculated leaves of Nicotiana benthamiana were examined, 7 dpi, under blue light, with visible light to outline leaf shape. 35S/FHV-C2 inoculated alone or with p19 silencing suppressor. FECT-eGFP is general high-expression positive control. F1DI (±p19) is a positive control for FHV/GFP expression and comprises FHV RNA1 plus a defective interfering construct of RNA2. All other inoculations included p19 unless otherwise mentioned. 2sg2KSS construct is included in the supplementary data. All other designations as in Fig. 2 (Color figure online)
To create a FHV vector competent for encapsidation by TMV CP, the TMV Oa was inserted into 35S/FHV-C2-GFP at two different positions (35S/FHV-C2-o1, o2), and adjacent to eGFP ORF. In order to test the influence of C-terminal TMV Oa fusion on eGFP expression, a third construct, 35S/FHV-C2-o3, was designed with the introduction of a stop codon at the natural stop site of eGFP, resulting in an eGFP fusion with B2 only at the N terminus. These three Oa-containing constructs (35S/FHV-C2-o1 to o3) were found to express only slightly less eGFP in leaves than the non-Oa, 35S/FHV-C2 (Figs. 3, 4a). This was unexpected since the TMV Oa sequence was added close to either the putative subgenomic promoter or the FHV 3′UTR. Little difference in fluorescence was observed between the three Oa-containing constructs.
eGFP expression in plants by FHV constructs and in 1-step and 2-step inoculation procedures. a eGFP fluorometry of N. benthamiana agroinoculated with various FHV constructs. p19, mock inoculation with p19 only; FECT-eGFP, high-expression positive control; 1-step and 2-step, co-agroinoculation or delayed TMV CP agroinoculation. Four replicates each treatment, except 15 replicates for 1-step and 2-step treatments. b eGFP expression compared in 1-step and 2-step agroinoculation procedures. FHV-C2-o3 without any FECT-TMV was also inoculated as a control (Color figure online)
To express the FHV silencing suppressor, B2 (Albariño et al. 2003), in conjunction with eGFP, we made variants of 35S/FHV-C4-2sg. To prevent the deletion of the eGFP ORF, we placed the eGFP ORF at the 3′ terminus of the virus, in contrast to the C4-2sg construct. Any homologous recombination between the two homologous subgenomic regions would delete B2 and Oa, but not eGFP, and deletion mutants would not be packaged as nanoparticles. It was observed that the 35S/FHV-C2-GFP construct clearly resulted in a brighter fluorescence than 35S/FHV-C4-2sg, which was further confirmed by fluorometry analysis (Fig. 4a).
To explore the impact of improved B2 expression, a portion of the FHV 3′UTR, which is normally downstream of the B2 ORF, was added to the internal B2 ORF followed by the TMV Oa. The final construct 35S/FHV2sg2 was created by adding 24 bp of C-terminal B2 sequence to aid eGFP expression by providing more natural context at the 3′ end of the ORF. We expected to see stronger eGFP expression and/or FHV B2 expression that would functionally replace p19. However, the FHV2sg2 vector did not significantly improve eGFP fluorescence expression compared with the original C4-2sg construct in planta (Fig. 3). Other constructs were built and tested, which included the precedent construct of FHV2sg2 (data not included) and a vector with the addition of strong Kozak context in pursuit of enhanced expression (FHV2sg2KSS, supplementary Fig. 1a). Neither resulted in any improvement in eGFP fluorescence (Fig. 3). In all cases, p19 was still required via co-agroinoculation for strong fluorescence. All subsequent experiments used the 35S/FHV-C2-o3 construct co-agroinoculated with p19.
Coexpression of FHV and FECT in Plants
To encapsidate FHV vector RNA in planta, a ratio of 20:1 mass ratio of TMV coat protein (CP) to RNA is required. The high-expression Foxtail mosaic virus vector, FECT [29], was used to produce TMV CP without being itself encapsidated. FECT produced TMV CP at a level comparable to the TMV vector JL24 [27], which expresses TMV CP as a native gene (Fig. 5).
Expression of TMV CP by FECT plant viral vector. Lane a FECT expressing TMV CP, Lane b TMV vector JL24 [23] expressing CP and eGFP. Both agroinoculations in N. benthamiana included p19 silencing suppressor. Far left lane protein marker (NEB # P7708) with sizes in kDa indicated
We next examined the ability of FHV and FECT to co-infect cells, to ensure that there was no replication interference. In a co-infection test system, 35S/FHV-C2 expressing eGFP and FECT expressing DsRed were co-agroinoculated into several leaves, resulting in a yellow-green fluorescence under blue light when viewed without magnification (Fig. 6a). In order to determine co-infection of single cells, co-infected leaves were reduced to protoplasts and the protoplasts were examined under a UV microscope. As seen in a representative photo (Fig. 6b), about 75 % of the eGFP positive cells are also DsRed positive, but not vice versa. FECT strongly infects the great majority of plant cells [29], as seen by the DsRed signals in Fig. 6b. FHV/eGFP is an insect virus construct and infects a much smaller number of plant cells, but those that are infected are mostly co-infected with FECT/DsRed, demonstrating the high frequency at which double infection occurs with this system, given the limitations of FHV infectivity itself.
Co-infection of plant cells by FHV and FECT viral vectors. a N. benthamiana plants were agroinoculated with p19 plus (left to right) 35S/FHVC2-o3/GFP, FECT/DsRed, or both vectors. b Protoplasts made from 4 dpi leaves co-agroinoculated with FHV-eGFP/FECT-DsRed (right leaf in (a)) were visualized for eGFP and DsRed fluorescence, showing that the majority of the FHV-eGFP infected cells were also infected with FECT (Color figure online)
eGFP Expression by FHV Enhanced by Delayed TMV CP Expression
Nanoparticles were produced by agroinoculation with 35S/p19, 35S/FHV-C2-o3, and 35S/FECT-TMV CP. The average size of our FHV-C2-o3 nanoparticles is estimated to be ~200nM based on the length of the FHV RNA genome (C2-o3; 4182 nts.), compared with wide-type TMV (6395 nts.) that generates a 300-nM particle (Fig. 7).
In all experiments, the presence of TMV CP at the time of FHV early infection (i.e., co-inoculation) led to reduced eGFP fluorescence. We hypothesized that CP binding the Oa early in infection impeded the replicative or translational events of FHV RNA. To test this, a “2-step” protocol was used in which the FECT/TMV CP inoculation was delivered 3 days after the FHV/GFP/Oa and p19 inoculations. The 2-step procedure consistently increased eGFP expression (Fig. 4).
Immune Response to Nanoparticles in Mice
In order to test the capacity of transencapsidated FHV RNA to express the eGFP transgene in mice, in planta transencapsidated FHV RNA was used to immunize BALB/c mice with in vitro transencapsidated FHV RNA or SFV RNA as encapsidation controls. Two doses of 15 or 30 μg encapsidated RNA (0.75 or 1.5 μg of RNA, respectively) were given by subcutaneous injection, without adjuvant. eGFP protein (15 μg) was used as a positive control, while PBS buffer was used as a negative control. Sera collected from mice before immunization and after a single dose were essentially negative for immune responses for all groups (data not shown). Weak but detectable anti-eGFP IgG responses were measured by ELISA after a second vaccination (pV2, Fig. 8), but all groups were statistically similar to PBS, including eGFP protein immunization. After a third immunization (pV3), all groups showed a strong trend toward augmented immunity against eGFP, but in large part were not significantly different than PBS, mainly due to high variance between responders and small group size. However, the highest dose of in planta encapsidated FHV (C2-o3, 30 µg) and eGFP protein control had IgG titers significantly higher than all in vitro encapsidated viral vector treatments. This confirmed the successful expression of eGFP transgene by transencapsidated FHV RNA, after uptake and presumed co-translational disassembly of TMV coat protein. This is notable, in light of low replication ability of FHV RdRp in animal cells at 37 degree [33].
In vivo analysis of FHV vaccine potency. Balb/C Mice (n = 3) were vaccinated 3 times, 2 weeks apart with indicated amounts (15 or 30 µg protein) of TMV-encapsidated FHV-eGFP, produced either in vitro by mixing RNA and coat protein, or in planta by coexpression of RNA and coat protein after agroinfiltration. PBS was used as a negative control, and in vitro encapsidated SFV-eGFP or 15 μg of eGFP protein was used as a positive control. ELISA analysis was used to determine anti-eGFP IgG titers on sera collected at 10 days after either vaccine 2 (pV2) or after vaccine 3 (pV3). Titers were measured against a known quantity of anti-eGFP standard (Vector labs) and shown as mean ± SEM using GraphPad Prism. Statistical analysis of differences between PBS and vaccine groups after vaccine 3 was evaluated by one-tailed t test. The asterisk indicates statistically significant difference from PBS control
Discussion
We have shown in this study that FHV can be encapsidated in planta with TMV coat protein and the resulting nanoparticle vaccines had improved characteristics compared to in vitro encapsidated FHV RNA. In previous studies, we demonstrated that SFV could be encapsidated in vitro with TMV coat protein [21]. TMV coat protein produced in vivo had also been used to assemble wild-type TMV virions in E. coli [34], and mRNAs had been encapsidated in planta to form TMV hybrid virions [35]. As well, Brome mosaic virus (BMV) RNA containing the TMV Oa was transencapsidated with TMV CP in barley protoplasts [36] and Rao and colleagues produced non-specific transencapsidated virions by coat protein of the similarly structured BMV in studying encapsidation specificity [37]. Though FHV virions use a multitude of molecular cues in virion assembly, similar to other icosahedral viruses [38], TMV and other tobamoviruses utilize a single Oa sequence to initiate assembly, with the remainder of the encapsidated sequence apparently without further molecular cues [39]. Thus, any RNA containing the TMV Oa should be able to be transencapsidated. It may be possible to extend this technique to other viral species for viral-vectored nanoparticle vaccine assembly in planta.
The individual components of the nanoparticles appeared to be produced at high levels. The FECT viral vector produced TMV CP at the same level as the native TMV vector, JL24 (Fig. 5). FHV vector levels, as measured by visually assessed fluorescence of eGFP (Fig. 3), were greater in side-by-side studies than the DI638 vector used in previous FHV work in N. benthamiana [37]. The coexpression of p19 silencing suppressor further boosted this eGFP expression even with FHV vector constructs that had an intact B2 silencing suppressor (Figs. 2, 3).
As a prerequisite for assembly, coexpression of both vectors in a single cell is necessary. The FECT vector was shown to express in the majority of cells harboring the FHV vector (Fig. 6). However, when FHV RNA and TMV CP vectors were co-inoculated, we saw a significant decrease in fluorescence. A supplementary experiment was performed in order to exclude the possibility of FECT interfering FHV replication (suppl. Fig. 2). This inhibition phenomenon is most likely mediated by classical coat protein resistance [40] and was previously observed by the Ahlquist group working with BMV transencapsidated by TMV CP. BMV RNAs 1 and 2 containing the TMV Oa decreased in replication 20-fold when co-inoculated with BMV RNA 3 expressing TMV CP [36]. This was theorized to be due to TMV CP binding to the BMV RNAs and interfering with replication. We investigated this hypothesis by separating the agroinoculation of FHV vector and TMV CP into two steps, delaying the expression of TMV CP until FHV RNA replication was sufficient to generate robust levels of eGFP protein. The two-step plants consistently showed higher expression of the viral eGFP transgene (Fig. 4), suggesting that RNA packaging by TMV CP reduced FHV RNA replication and/or translation.
Several modifications were made in an attempt to improve FHV vector replication in plants. The addition of TMV Oa led to strong inhibition of BMV RNA replication even in the absence of TMV CP in a previous study [36]. However, we observed only a slight decrease in eGFP production by the FHV vectors carrying Oa. C2 constructs carrying Oa at two different sites (C2-o1 and C2-o2) did not differ significantly in eGFP fluorescence produced. Recreating a native C-terminus for eGFP (C2-o3) also had no effect. Constructs with unmodified B2 silencing suppressor ORFs (2sg2 series) were less effective than the C2 series with the B2 ORF fused to eGFP. These were longer constructs, but the shorter C4 construct was also less fluorescent in mammalian cells than the C2 construct (Fig. 1b), suggesting the common C4 insertion site as detrimental. Ultimately, the inclusion of p19 as a co-inoculant was the sole factor in achieving high eGFP expression in plants from the FHV vectors, re-confirming the importance of mitigating RNA silencing in planta.
It is possible that the size of the duplicated subgenomic promoter in the 2sg2 series was insufficient, since a longer FHV subgenomic promoter segment was found more efficacious in a previous study [41]. Beyond the core nts. 2518–2777, the region from nt. 2302–2518 may serve as an important enhancer [42]. Polarity preference was found on FHV [41] and other positive-strand RNA viruses; specifically, that two pieces of sgRNA were replicated at different levels, with the longer one (closer to replicase) being dominant. This may explain why we see more eGFP fluorescence in the 35S/FHV-C4-2sg, which has the eGFP ORF included in the first sgRNA3, than in the FHV-2sg2 series, which has eGFP ORF included in the second sgRNA3.
In order to verify the capacity of these transencapsidated nanoparticles to express transgene in animal cells, FHV C2-o3-encapsidated particles were used as a vaccine, and an IgG antibody response to eGFP was measured (Fig. 8). Despite the reported deficiency of FHV replicase to function well in 37 degree [33], a titer of anti-eGFP antibody equal to that of 15 μg eGFP protein was observed after three injections with 30 μg of FHV C2-o3 nanoparticles (1.5 μg FHV RNA). This demonstrated delivery and expression of the eGFP transgene and suggested a considerable boosting of the immune response by RNA antigen delivery. In vitro generated FHV and SFV/TMV CP nanoparticles produced a significantly lower immune response in this study, possibly due to lower percentage of 5′ capping, which is known to affect translation efficiency.
During nanoparticle in planta assembly, FHV subgenomic RNA3 may also be encapsidated by TMV coat proteins, co-purified and be represented in mice injections. The possibility of sgRNA3 being used as mRNA templates has been considered, since sgRNA3 also contains eGFP sequence and contains a TMV Oa. However, from the numerous TEM images, it is apparent that the amount of sgRNA3 nanoparticles (~67nM) is not evident or a minority of the particles, and the majority nanoparticles are of full length (200 nm). Furthermore, from the previous literature, the non-replicating mRNA vaccination strategy has largely relied on extensive chemical modifications, additional use of adjuvants [43], and an ex vivo route to transfect dendritic cells [44]. In two studies using eGFP mRNA to transfect dendritic cells, eGFP either degraded too rapidly due to the lack of additional targeting signals [45], or was expressed well in dendritic cells but failed to trigger dendritic cell maturation without using inducing agents [46]. Overall, it is more likely that a functional replicase and a self-replicating viral RNA account for the bulk of the immune stimulation observed in this work, rather than translation from sgRNA3.
Several improvements can be made to the utility of TMV coat encapsidated RNA. In order to increase immune activation and greater CD4 T cell response, future optimization may include the use of other viruses with a replicase active at 37 degree, such as Nodamura virus [33]. Peptide-directed endosomal escape of nanoparticles [47–49] may also increase animal cell infection and subsequent protein accumulation. In our study, eGFP was used to track viral expression of eGFP in plant and animal cells. Expression of a more potent immunogen (e.g., ovalbumin) with better characterized antigenicity should also improve both antibody and T cell immunogenicity after nanoparticle vaccination.
In conclusion, we were able to produce FHV RNA and TMV CP, in the same plant cell, resulting in assembly of rod-shaped packaged RNA. These in planta-produced nanoparticles were shown to induce an antigen-specific immunogenicity exceeding that of in vitro-packaged RNA nanoparticles. Our next tasks are to investigate cellular localization in planta of FHV RNA and TMV CP and to optimize heterologous virion assembly. We will also seek to target the hybrid virion nanoparticles to the correct compartment in the mammalian cell in order to facilitate TMV CP virion disassembly and improved RNA 1 replication. Completion of these goals will answer basic virological questions of component trafficking, disassembly, and replication in the process of optimizing vaccine production and potency.
References
Gutierro, I., Hernandez, R. M., Igartua, M., Gascon, A. R., & Pedraz, J. L. (2002). Influence of dose and immunization route on the serum Ig G antibody response to BSA loaded PLGA microspheres. Vaccine, 20, 2181–2190.
Gutierro, I., Hernandez, R. M., Igartua, M., Gascon, A. R., & Pedraz, J. L. (2002). Size dependent immune response after subcutaneous, oral and intranasal administration of BSA loaded nanospheres. Vaccine, 21, 67–77.
Dobrovolskaia, M. A., & McNeil, S. E. (2007). Immunological properties of engineered nanomaterials. Nature Nanotechnology, 2, 469–478.
McCormick, A. A., Reddy, S., Reinl, S. J., Cameron, T. I., Czerwinkski, D. K., Vojdani, F., et al. (2008). Plant-produced idiotype vaccines for the treatment of non-Hodgkin’s lymphoma: safety and immunogenicity in a phase I clinical study. Proceedings of the National Academy of Sciences of the United States of America, 105, 10131–10136.
Aires, K. A., Cianciarullo, A. M., Carneiro, S. M., Villa, L. L., Boccardo, E., Perez-Martinez, G., et al. (2006). Production of human papillomavirus type 16 L1 virus-like particles by recombinant Lactobacillus casei cells. Applied and Environmental Microbiology, 72, 745–752.
Huang, Z., Santi, L., LePore, K., Kilbourne, J., Arntzen, C. J., & Mason, H. S. (2006). Rapid, high-level production of hepatitis B core antigen in plant leaf and its immunogenicity in mice. Vaccine, 24, 2506–2513.
Zhou, J., Sun, X. Y., Stenzel, D. J., & Frazer, I. H. (1991). Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology, 185, 251–257.
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S., & Reis e Sousa, C. (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science, 303, 1529–1531.
Lund, J. M., Alexopoulou, L., Sato, A., Karow, M., Adams, N. C., Gale, N. W., et al. (2004). Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proceedings of the National Academy of Sciences of the United States of America, 101, 5598–5603.
Schwarz, K., Storni, T., Manolova, V., Didierlaurent, A., Sirard, J. C., Rothlisberger, P., & Bachmann, M. F. (2003). Role of Toll-like receptors in costimulating cytotoxic T cell responses. European Journal of Immunology, 33, 1465–1470.
Pogue, G. P., Lindbo, J. A., Garger, S. J., & Fitzmaurice, W. P. (2002). Making an ally from an enemy: plant virology and the new agriculture. Annual Review of Phytopathology, 40, 45–74.
Liu, R., Vaishnav, R. A., Roberts, A. M., & Friedland, R. P. (2013). Humans have antibodies against a plant virus: Evidence from tobacco mosaic virus. PLoS One, 8, e60621.
McCormick, A. A., Corbo, T. A., Wykoff-Clary, S., Palmer, K. E., & Pogue, G. P. (2006). Chemical conjugate TMV-peptide bivalent fusion vaccines improve cellular immunity and tumor protection. Bioconjugate Chemistry, 17, 1330–1338.
McCormick, A. A., Corbo, T. A., Wykoff-Clary, S., Nguyen, L. V., Smith, M. L., Palmer, K. E., & Pogue, G. P. (2006). TMV-peptide fusion vaccines induce cell-mediated immune responses and tumor protection in two murine models. Vaccine, 24, 6414–6423.
Smith, M. L., Lindbo, J. A., Dillard-Telm, S., Brosio, P. M., Lasnik, A. B., McCormick, A. A., et al. (2006). Modified tobacco mosaic virus particles as scaffolds for display of protein antigens for vaccine applications. Virology, 348, 475–488.
Mallajosyula, J. K., Hiatt, E., Hume, S., Johnson, A., Jeevan, T., Chikwamba, R., et al. (2013). Single-dose monomeric HA subunit vaccine generates full protection from influenza challenge. Human Vaccines & Immunotherapeutics, 10, 586–595.
Fraile, A., Escriu, F., Aranda, M. A., Malpica, J. M., Gibbs, A. J., & Garcia-Arenal, F. (1997). A century of tobamovirus evolution in an Australian population of Nicotiana glauca. Journal of Virology, 71, 8316–8320.
Kemnade, J. O., Seethammagari, M., Collinson-Pautz, M., Kaur, H., Spencer, D. M., & McCormick, A. A. (2014). Tobacco mosaic virus efficiently targets DC uptake, activation and antigen-specific T cell responses in vivo. Vaccine, 32, 4228–4233.
Koo, M., Bendahmane, M., Lettieri, G. A., Paoletti, A. D., Lane, T. E., Fitchen, J. H., et al. (1999). Protective immunity against murine hepatitis virus (MHV) induced by intranasal or subcutaneous administration of hybrids of tobacco mosaic virus that carries an MHV epitope. Proceedings of the National Academy of Sciences of the United States of America, 96, 7774–7779.
Yin, Z., Nguyen, H. G., Chowdhury, S., Bentley, P., Bruckman, M. A., Miermont, A., et al. (2012). Tobacco mosaic virus as a new carrier for tumor associated carbohydrate antigens. Bioconjugate Chemistry, 23, 1694–1703.
Smith, M. L., Corbo, T., Bernales, J., Lindbo, J. A., Pogue, G. P., Palmer, K. E., & McCormick, A. A. (2007). Assembly of trans-encapsidated recombinant viral vectors engineered from Tobacco mosaic virus and Semliki Forest virus and their evaluation as immunogens. Virology, 358, 321–333.
Lundstrom, K. (2003). Semliki Forest virus vectors for gene therapy. Expert Opinion on Biological Therapy, 3, 771–777.
Mahraj, P. D., Mallajosyula, J. K., Lee, G., Thi, P., Zhou, Y., Kearney, C. M. & McCormick, A. A. (2014). Nanoparticle encapsidation of Flock House virus by auto assembly of Tobacco Mosaic virus coat protein. International Journal of Molecular Sciences, 15, 18540–18556.
Johnson, K. L., & Ball, L. A. (1999). Induction and maintenance of autonomous flock house virus RNA1 replication. Journal of Virology, 73, 7933–7942.
Selling, B. H., Allison, R. F., & Kaesberg, P. (1990). Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proceedings of the National Academy of Sciences of the United States of America, 87, 434–438.
Price, B., Roeder, M., & Ahlquist, P. (2000). DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. Journal of Virology, 74, 11724–11733.
Lindbo, J. A. (2007). High-efficiency protein expression in plants from agroinfection-compatible Tobacco mosaic virus expression vectors. BMC Biotechnology, 7, 52.
Turner, D. R., & Butler, P. J. (1986). Essential features of the assembly origin of tobacco mosaic virus RNA as studied by directed mutagenesis. Nucleic Acids Research, 14, 9229–9242.
Liu, Z., & Kearney, C. M. (2010). An efficient Foxtail mosaic virus vector system with reduced environmental risk. BMC Biotechnology, 10, 88.
Dasgupta, R., Cheng, L. L., Bartholomay, L. C., & Christensen, B. M. (2003). Flock house virus replicates and expresses green fluorescent protein in mosquitoes. Journal of General Virology, 84, 1789–1797.
Price, B. D., Ahlquist, P., & Ball, L. A. (2002). DNA-directed expression of an animal virus RNA for replication-dependent colony formation in Saccharomyces cerevisiae. Journal of Virology, 76, 1610–1616.
Scholthof, H. B. (2006). The Tombusvirus-encoded P19: From irrelevance to elegance. Nature Reviews Microbiology, 4, 405–411.
Ball, L. A., Amann, J. M., & Garrett, B. K. (1992). Replication of nodamura virus after transfection of viral-RNA into mammalian-cells in culture. Journal of Virology, 66, 2326–2334.
Hwang, D. J., Roberts, I. M., & Wilson, T. M. (1994). Expression of tobacco mosaic virus coat protein and assembly of pseudovirus particles in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 91, 9067–9071.
Sleat, D. E., Gallie, D. R., Watts, J. W., Deom, C. M., Turner, P. C., Beachy, R. N., & Wilson, T. M. (1988). Selective recovery of foreign gene transcripts as virus-like particles in TMV-infected transgenic tobaccos. Nucleic Acids Research, 16, 3127–3140.
Sacher, R., French, R., & Ahlquist, P. (1988). Hybrid brome mosaic virus RNAs express and are packaged in tobacco mosaic virus coat protein in vivo. Virology, 167, 15–24.
Annamalai, P., Rofail, F., Demason, D. A., & Rao, A. L. (2008). Replication-coupled packaging mechanism in positive-strand RNA viruses: Synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in Nicotiana benthamiana cells by agroinfiltration. Journal of Virology, 82, 1484–1495.
Rao, A. L. (2006). Genome packaging by spherical plant RNA viruses. Annual Review of Phytopathology, 44, 61–87.
Wilson, T. M., & McNicol, J. W. (1995). A conserved, precise RNA encapsidation pattern in Tobamovirus particles. Archives of Virology, 140, 1677–1685.
Abel, P. P., Nelson, R. S., De, B., Hoffmann, N., Rogers, S. G., Fraley, R. T., & Beachy, R. N. (1986). Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science, 232, 738–743.
Lindenbach, B. D., Sgro, J. Y., & Ahlquist, P. (2002). Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. Journal of Virology, 76, 3905–3919.
Sztuba-Solinska, J., Stollar, V., & Bujarski, J. J. (2011). Subgenomic messenger RNAs: Mastering regulation of (+)-strand RNA virus life cycle. Virology, 412, 245–255.
Phua, K. K., Nair, S. K., & Leong, K. W. (2014). Messenger RNA (mRNA) nanoparticle tumour vaccination. Nanoscale, 6, 7715–7729.
Weiner, D. B. (2013). RNA-based vaccination: Sending a strong message. Molecular Therapy, 21, 506–508.
Bonehill, A., Heirman, C., Tuyaerts, S., Michiels, A., Breckpot, K., Brasseur, F., et al. (2004). Messenger RNA-electroporated dendritic cells presenting MAGE-A3 simultaneously in HLA class I and class II molecules. The Journal of Immunology, 172, 6649–6657.
Van Tendeloo, V. F., Ponsaerts, P., Lardon, F., Nijs, G., Lenjou, M., Van Broeckhoven, C., et al. (2001). Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: Superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood, 98, 49–56.
Tkachenko, A. G., Xie, H., Coleman, D., Glomm, W., Ryan, J., Anderson, M. F., et al. (2003). Multifunctional gold nanoparticle-peptide complexes for nuclear targeting. Journal of the American Chemical Society, 125, 4700–4701.
Oliveira, S., van Rooy, I., Kranenburg, O., Storm, G., & Schiffelers, R. M. (2007). Fusogenic peptides enhance endosomal escape improving siRNA-induced silencing of oncogenes. International Journal of Pharmaceutics, 331, 211–214.
Erazo-Oliveras, A., Muthukrishnan, N., Baker, R., Wang, T.-Y., & Pellois, J.-P. (2012). Improving the endosomal escape of cell-penetrating peptides and their cargos: Strategies and challenges. Pharmaceuticals, 5, 1177–1209.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, Y., Maharaj, P.D., Mallajosyula, J.K. et al. In planta Production of Flock House Virus Transencapsidated RNA and Its Potential Use as a Vaccine. Mol Biotechnol 57, 325–336 (2015). https://doi.org/10.1007/s12033-014-9826-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-014-9826-1