Abstract
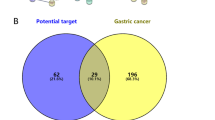
The Fucaceae family of marine brown algae includes Ascophyllum nodosum. Fucosterol (FSL) is a unique bioactive component that was identified through GC-MS analysis of the hydroalcoholic extract of A. nodosum. Fucosterol's mechanism of action towards hepatocellular cancer was clarified using network pharmacology and docking study techniques. The probable target gene of FSL has been predicted using the TargetNet and SwissTargetPred databases. GeneCards and the DisGNet database were used to check the targeted genes of FSL. By using the web programme Venny 2.1, the overlaps of FSL and HCC disease demonstrated that 18 genes (1.3%) were obtained as targeted genes Via the STRING database, a protein–protein interaction (PPI) network with 18 common target genes was constructed. With the aid of CytoNCA, hub genes were screened using the Cytoscape software, and the targets' hub genes were exported into the ShinyGo online tool for study of KEGG and gene ontology enrichment. Using the software AutoDock, a hub gene molecular docking study was performed. Ten genes, including AR, CYP19A1, ESR1, ESR2, TNF, PPARA, PPARG, HMGCR, SRC, and IGF1R, were obtained. The 10 targeted hubs docked with FSL successfully. The active components FSL of ASD, the FSL, are engaged in fatty liver disease, cancer pathways, and other signalling pathways, which could prove beneficial for the management of HCC.







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from public databases.
References
Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. https://doi.org/10.1038/s41571-018-0073-4.
Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019. https://doi.org/10.1056/NEJMra1713263.
Neophytou CM, Panagi M, Stylianopoulos T, Papageorgis P. The role of tumor microenvironment in cancer metastasis: molecular mechanisms and therapeutic opportunities. Cancers. 2021;13(9):2053. https://doi.org/10.3390/cancers13092053.
Le Grazie M, Biagini MR, Tarocchi M, Polvani S, Galli A. Chemotherapy for hepatocellular carcinoma: the present and the future. World J Hepatol. 2017;9(21):907. https://doi.org/10.4254/wjh.v9.i21.907.
Raza A, Sood GK. Hepatocellular carcinoma review: current treatment and evidence-based medicine. World J Gastroenterol. 2014;20(15):4115–27. https://doi.org/10.3748/wjg.v20.i15.4115.
Chen CP. Role of radiotherapy in the treatment of hepatocellular carcinoma. J Clin Transl Hepatol. 2019;7(2):183. https://doi.org/10.14218/JCTH.2018.00060.
Liu JKH, Irvine AF, Jones RL, Samson A. Immunotherapies for hepatocellular carcinoma. Cancer Med. 2022;11(3):571–91. https://doi.org/10.1002/cam4.4468.
Xiang Y, Guo Z, Zhu P, Chen J, Huang Y. Traditional Chinese medicine as a cancer treatment: modern perspectives of ancient but advanced science. Cancer Med. 2019;8(5):1958–75. https://doi.org/10.1002/cam4.2108.
Ugarte RA, Craigie JS, Critchley AT. Fucoid flora of the rocky intertidal of the Canadian Maritimes: implications for the future with rapid climate change. Seaweeds Role Glob Chang Environ. 2010. https://doi.org/10.1007/978-90-481-8569-6_5.
Gull N, Arshad F, Naikoo GA. Recent advances in anticancer activity of novel plant extracts and compounds from curcuma longa in hepatocellular carcinoma. J Gastrointest Cancer. 2022. https://doi.org/10.1007/s12029-022-00809-z.
Iqbal J, Abbasi BA, Ahmad R, Mahmood T, Kanwal S, Ali B, Khalil AT, Shah SA, Alam MM, Badshah H. Ursolic acid a promising candidate in the therapeutics of breast cancer: current status and future implications. Biomed Pharmacother. 2018;108:752 6. https://doi.org/10.1016/j.biopha.2018.09.096.
Agarwal G, Carcache PJB, Addo EM, Kinghorn AD. Current status and contemporary approaches to the discovery of antitumor agents from higher plants. Biotechnol Adv. 2020;38:107337. https://doi.org/10.1016/j.biotechadv.2019.01.004.
Iqbal J, Abbasi BA, Batool R, Mahmood T, Ali B, Khalil AT, Kanwal S, Shah SA, Ahmad R. Potential phytocompounds for developing breast cancer therapeutics: nature’s healing touch. Eur J Pharmacol. 2018;827:125–48. https://doi.org/10.1016/j.ejphar.2018.03.007.
Avtanski D, Poretsky L. Phyto-polyphenols as potential inhibitors of breast cancer metastasis. Mol Med. 2018;24(1):1–17. https://doi.org/10.1186/s10020-018-0032-7.
Shehzad A, Qureshi M, Anwar MN, Lee YS. Multifunctional curcumin mediate multitherapeutic effects. J Food Sci. 2017;82(9):2006–15. https://doi.org/10.1111/1750-3841.13793.
Pan Z, Zhuang J, Ji C, Cai Z, Liao W, Huang Z. Curcumin inhibits hepatocellular carcinoma growth by targeting VEGF expression. Oncol Lett. 2018;15(4):4821–6. https://doi.org/10.3892/ol.2018.7988.
Xu MX, Zhao L, Deng C, Yang LU, Wang Y, Guo T, Li L, Lin J, Zhang L. Curcumin suppresses proliferation and induces apoptosis of human hepatocellular carcinoma cells via the wnt signaling pathway. Int J Oncol. 2013;43(6):1951–9. https://doi.org/10.3892/ijo.2013.2107.
Teng CF, Yu CH, Chang HY, Hsieh WC, Wu TH, Lin JH, Wu HC, Jeng LB, Su IJ. Chemopreventive effect of phytosomal curcumin on hepatitis B virus-related hepatocellular carcinoma in a transgenic mouse model. Sci Rep. 2019;9(1):10338. https://doi.org/10.1038/s41598-019-46891-5.
Matias D, Rijo P, Pinto RC. Phytosomes as biocompatible carriers of natural drugs. Curr Med Chem. 2017;24(6):568–89. https://doi.org/10.2174/0929867323666161028160855.
Mirzaei H, Shakeri A, Rashidi B, Jalili A, Banikazemi Z, Sahebkar A. Phytosomal curcumin: a review of pharmacokinetic, experimental and clinical studies. Biomed Pharmacother. 2017;129:101 7. https://doi.org/10.1016/j.carbpol.2015.04.057.
Yuan Y, Macquarrie D. Microwave assisted extraction of sulfated polysaccharides (fucoidan) from Ascophyllum nodosum and its antioxidant activity. Carbohydr Polym. 2015;129:101 7. https://doi.org/10.1016/j.carbpol.2015.04.057.
Venardou B, O’Doherty JV, Garcia-Vaquero M, Kiely C, Rajauria G, McDonnell MJ, Ryan MT, Sweeney T. Evaluation of the antibacterial and prebiotic potential of Ascophyllum nodosum and its extracts using selected bacterial members of the pig gastrointestinal microbiota. Mar Drugs. 2021;20(1):41. https://doi.org/10.3390/md20010041.
Ajah HA. In vitro and in vivo studies on the antifungal activity of probiotics and seaweed extract (Ascophyllum nodosum). Int J Innov Sci Eng Technol. 2016;3(4):306–12.
Chauhan BS, Kumar R, Kumar P, Kumar P, Sinha S, Mishra SK, Tiwari KN, Critchley AT, Prithiviraj B, Srikrishna S. Neuroprotective potential of flavonoid rich Ascophyllum nodosum (FRAN) fraction from the brown seaweed on an Aβ42 induced Alzheimer’s model of Drosophila. Phytomed. 2022;95:153872. https://doi.org/10.1016/j.phymed.2021.153872.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;42(W1):W32–8. https://doi.org/10.1093/nar/gkz382.
Zhi-Jiang Y, Jie D, Yu-Jing C, Min-Feng Z, Ming W, Ning-Ning W, Shan W, Ai-Ping Lu, Cao D-S. TargetNet: a web service for predicting potential drug-target interaction profiling via multi-target SAR models. J Comput Aided Mol Des. 2016;30:413–24. https://doi.org/10.1007/s10822-016-9915-2.
Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota-Madi A. GeneCards version 3: the human gene integrator. Database. 2010. https://doi.org/10.1093/database/baq020.
Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET cytoscape app: exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021;19(7789):2960–7. https://doi.org/10.1016/j.csbj.2021.05.015.
Lotia S, Montojo J, Dong Y, Bader GD, Pico AR. Cytoscape app store. Bioinformatics. 2013;29(10):1350–1. https://doi.org/10.1093/bioinformatics/btt138.
Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36(8):2628–9. https://doi.org/10.1093/bioinformatics/btz931.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019. https://doi.org/10.1093/bioinformatics/btz931.
Desai AG, Qazi GN, Ganju RK, El-Tamer M, Singh J, Saxena AK, Bedi YS, Taneja SC, Bhat HK. Medicinal plants and cancer chemoprevention. Curr Drug Metab. 2008;9(7):581–91. https://doi.org/10.2174/138920008785821657.
Meinita MDN, Harwanto D, Tirtawijaya G, Negara BFSP, Sohn JH, Kim JS, Choi JS. Fucosterol of marine macroalgae: bioactivity, safety and toxicity on organism. Mar Drugs. 2021;19(10):545. https://doi.org/10.3390/md19100545.
Liang J, Lv J, Liu Z. Identification of dysfunctional biological pathways and their synergistic mechanism in hepatocellular carcinoma process. Exp Mol Pathol. 2015;98(3):540–5. https://doi.org/10.1016/j.yexmp.2015.03.028.
Hishida M, Nomoto S, Inokawa Y, Hayashi M, Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S, Nakayama G, Fujii T, Sugimoto H, Koike M, Fujiwara M, Takeda S, Kodera Y. Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Oncol. 2013;43(1):88–94. https://doi.org/10.3892/ijo.2013.1951.
Villa E, Moles A, Ferretti I, Buttafoco P, Grottola A, Buono M, De Santis M, Manenti F. Natural history of inoperable hepatocellular carcinoma: estrogen receptors’ status in the tumor is the strongest prognostic factor for survival. Hepatology. 2000;32(2):233–8. https://doi.org/10.1053/jhep.2000.9603.
Kocanova S, Mazaheri M, Caze-Subra S, Bystricky K. Ligands specify estrogen receptor alpha nuclear localization and degradation. BMC Cell Biol. 2010;11:98. https://doi.org/10.1186/1471-2121-11-98.
Ochiai I, Matsuda K, Nishi M, Ozawa H, Kawata M. Imaging analysis of subcellular correlation of androgen receptor and estrogen receptor alpha in single living cells using green fluorescent protein color variants. Mol Endocrinol. 2004;18(1):26–42. https://doi.org/10.1210/me.2002-0262.
Helsen C, Kerkhofs S, Clinckemalie L, Spans L, Laurent M, Boonen S, Vanderschueren D, Claessens F. Structural basis for nuclear hormone receptor DNA binding. Mol Cell Endocrinol. 2012;348(2):411–7. https://doi.org/10.1016/j.mce.2011.07.025.
Lonard DM, O’Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125(3):411–4. https://doi.org/10.1016/j.cell.2006.04.021.
Gadaleta RM, Magnani L. Nuclear receptors and chromatin: an inducible couple. J Mol Endocrinol. 2014;52(2):R137–49. https://doi.org/10.1530/JME-13-0170.
Kininis M, Kraus WL. A global view of transcriptional regulation by nuclear receptors: gene expression, factor localization, and dna sequence analysis. Nucl Recept Signal. 2008;6:e005. https://doi.org/10.1621/nrs.06005.
Clocchiatti A, Cora E, Zhang Y, Dotto GP. Sexual dimorphism in cancer. Nat Rev Cancer. 2016;16(5):330–9. https://doi.org/10.1038/nrc.2016.30.
Tiegs G, Horst AK. TNF in the liver: targeting a central player in inflammation. Semin Immunopathol. 2022;44:445–59. https://doi.org/10.1007/s00281-022-00910-2.
Chang C, Lee SO, Yeh S, Chang TM. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene. 2014;33(25):3225–34. https://doi.org/10.1038/onc.2013.274.
Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20(13):3001–15. https://doi.org/10.1200/JCO.2002.10.018.
Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25(2):276–308. https://doi.org/10.1210/er.2002-0032.
Yoon G, Kim JY, Choi YK, Won YS, Lim IK. Direct activation of TGF-beta1 transcription by androgen and androgen receptor complex in Huh7 human hepatoma cells and its tumor in nude mice. J Cell Biochem. 2006;97(2):393–411. https://doi.org/10.1002/jcb.20638.
Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21(16):2005–17. https://doi.org/10.1101/gad.1564207.
Jiang X, Kanda T, Nakamoto S, Miyamura T, Wu S, Yokosuka O. Involvement of androgen receptor and glucose-regulated protein 78 kDa in human hepatocarcinogenesis. Exp Cell Res. 2014;323(2):326–36. https://doi.org/10.1016/j.yexcr.2014.02.017.
Nagasue N, Ito A, Yukaya H, Ogawa Y. Androgen receptors in hepatocellular carcinoma and surrounding parenchyma. Gastroenterology. 1985;89(3):643–7. https://doi.org/10.1016/0016-5085(85)90463-9.
Kaewlert W, Sakonsinsiri C, Namwat N, Sawanyawisuth K, Ungarreevittaya P, Khuntikeo N, Armartmuntree N, Thanan R. The importance of CYP19A1 in estrogen receptor-positive cholangiocarcinoma. Horm Canc. 2018;9(6):408–19. https://doi.org/10.1007/s12672-018-0349-2.
Ngo MHT, Jeng HY, Kuo YC, Nanda JD, Brahmadhi A, Ling TY, Chang TS, Huang YH. The role of IGF/IGF-1R signaling in hepatocellular carcinomas: stemness-related properties and drug resistance. Int J Mol Sci. 2021;22(4):1931. https://doi.org/10.3390/ijms22041931.
Wu L, Guo C, Wu J. Therapeutic potential of PPARγ natural agonists in liver diseases. J Cell Mol Med. 2020;24(5):2736–48. https://doi.org/10.1111/jcmm.15028.
Zhang S, Yu D. Targeting Src family kinases in anti-cancer therapies: turning promise into triumph. Trends Pharmacol Sci. 2012;33(3):122–8. https://doi.org/10.1016/j.tips.2011.11.002.
Kumar P, Singh AK, Verma P, Tiwari KN, Mishra SK. Network pharmacology-based study on apigenin present in the methanolic fraction of leaves extract of Cestrum nocturnum L. to uncover mechanism of action on hepatocellular carcinoma. Med Oncol. 2022. https://doi.org/10.1007/s12032-022-01759-z.
Kumar P, Singh AK, Tiwari KN, et al. Identification and validation of hub genes as promising diagnostic signature in hepatocellular carcinoma based on integrated bioinformatics approach. Sci Rep. 2022. https://doi.org/10.1038/s41598-022-22059-6.
Van Diepen JA, Jansen PA, Ballak DB, Hijmans A, Hooiveld GJ, Rommelaere S, Galland F, Naquet P, Rutjes FP, Mensink RP, Schrauwen P, Tack CJ, Netea MG, Kersten S, Schalkwijk J, Stienstra R. PPAR-alpha dependent regulation of vanin-1 mediates hepatic lipid metabolism. J Hepatol. 2014;61(2):366–72. https://doi.org/10.1016/j.jhep.2014.04.013.
Palmer CNA, Hsu M-H, Griffin KJ, Johnson EF. Novel sequence determinants in peroxisome proliferator signaling. J Biol Chem. 1995;270(27):16114–21. https://doi.org/10.1074/jbc.270.27.16114.
Cheng B, Li T, Li F. Use of network pharmacology to investigate the mechanism by which allicin ameliorates lipid metabolism disorder in hepg2 cells. Evid Based Complement Alternat Med. 2021;2021(12):3956504. https://doi.org/10.1155/2021/3956504.
Berger A, Monnard I, Baur M, Charbonnet C, Safonova I, Jomard A. Epidermal anti-inflammatory properties of 5,11,14 20:3: effects on mouse ear edema, PGE2 levels in cultured keratinocytes, and PPAR activation. Lipids Health Dis. 2002;1(1):5. https://doi.org/10.1186/1476-511x-1-5.
Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Progress Lipid Res. 2006;45(2):120–59. https://doi.org/10.1016/j.plipres.2005.12.002.
Tan Y, Wang M, Yang K, Chi T, Liao Z, Wei P. PPAR-α modulators as current and potential cancer treatments. Front Oncol. 2021;23(11):599995. https://doi.org/10.3389/fonc.2021.599995.
Acknowledgements
The author Kajal Singh gratefully to acknowledge to Department of pharmaceutical engineering and Technology, IIT, BHU, Department of Botany, MMV, BHU, and Galgotias University, Greater Noida, Uttar Pradesh for their immense support. Author Amit Kumar Singh acknowledge the MHRD, New Delhi for Funding support. Author Pradeep Kumar Highly thankful to University Grant Commission, New Delhi for providing Rajiv Gandhi National Fellowship.
Funding
The research involved no particular grants either public, commercial, or non-profit financing entities.
Author information
Authors and Affiliations
Contributions
KS, PK, and AKS designed the concept of work, NS, SS, SA, and RD: Data analysis, interpretation of results, and manuscript preparation, SA, RD, SS, and NS: extract preparation and data interpretation, SKM and KNT, AKT, BR, and AS: designed the work and reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest in between author.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Research involving human participants and/or animals
This study does not contain any studies with human participants or animals performed by the author.
Consent to participant
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, K., Kumar, P., Singh, A.K. et al. In silico and network pharmacology analysis of fucosterol: a potent anticancer bioactive compound against HCC. Med Oncol 41, 130 (2024). https://doi.org/10.1007/s12032-024-02374-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-024-02374-w