Abstract
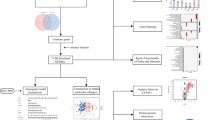
Lung cancer is one of the world’s most common and deadly cancers. The two main types of lung cancer are non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). More than 85% of lung cancers are NSCLC. Genetic factors play a significant role in the risk of NSCLC. Growing studies focus on studying risk factors at the molecular level. The aim of the study is to build a pipeline to integrate Genome-wide association analysis (GWAS) and transcriptomics data with machine learning to effectively identify genetic risk factors of NSCLC. GWAS datasets and GWAS summary data were downloaded from GWAS catalog, which include lung carcinoma genetic variants among the European population. Then, with the GWAS summary, data functional analysis of significant SNPs was performed using a webserver called FUMAGWAS. The transcriptomics data of NSCLC and non-NSCLC people were used to build a machine learning model to identify the key genes that help predict the NSCLC. The top up-regulation and down-regulation genes were identified by the BART cancer webserver, and the mechanistic roles of the genes were validated by literature review. By performing integrative analysis of GWAS and transcriptomics analysis using machine learning, we identified multiple SNPs and genes that related to NSCLC. The computational pipeline may facilitate the biomarker discovery for NSCLC and other diseases.




Similar content being viewed by others
References
Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30.
Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16:e165-172.
Skřičková J, Kadlec B, Venclíček O, Merta Z. Lung cancer. Cas Lek Cesk. 2018;157:226–36.
Webb JL, Burns RE, Brown HM, LeRoy BE, Kosarek CE. Squamous cell carcinoma. Compend Contin Educ Vet. 2009;31:E9.
Ettinger DS, et al. Non-small cell lung cancer, version 2. 2013. J Natl Compr Canc Netw. 2013;11:645–53.
Sinha R, et al. Fried, well-done red meat and risk of lung cancer in women (United States). Cancer Causes Control CCC. 1998;9:621–30.
Ebrahimi H, et al. Global, regional, and national burden of respiratory tract cancers and associated risk factors from 1990 to 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Respir Med. 2021;9:1030–49.
Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41:1–24.
Loeb LA, Ernster VL, Warner KE, Abbotts J, Laszlo J. Smoking and lung cancer: an overview. Cancer Res. 1984;44:5940–58.
Tammemägi MC, Berg CD, Riley TL, Cunningham CR, Taylor KL. Impact of lung cancer screening results on smoking cessation. J Natl Cancer Inst. 2014;106:dju084.
Doll R, Peto R, Boreham J, Sutherland I. Mortality from cancer in relation to smoking: 50 years observations on British doctors. Br J Cancer. 2005;92:426–9.
Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J. 2016;48:889–902.
Lorenzo-González M, et al. Radon exposure: a major cause of lung cancer. Expert Rev Respir Med. 2019;13:839–50.
Lee SS, Cheah YK. The interplay between MicroRNAs and cellular components of tumour microenvironment (TME) on Non-small-cell lung cancer (NSCLC) progression. J Immunol Res. 2019;2019:3046379.
Altorki NK, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19:9–31.
Hsieh C-H, et al. An innovative NRF2 nano-modulator induces lung cancer ferroptosis and elicits an immunostimulatory tumor microenvironment. Theranostics. 2021;11:7072–91.
Wu J, et al. A risk model developed based on tumor microenvironment predicts overall survival and associates with tumor immunity of patients with lung adenocarcinoma. Oncogene. 2021;40:4413–24.
Romaszko AM, Doboszyńska A. Multiple primary lung cancer: a literature review. Adv Clin Exp Med Off Organ Wroclaw Med Univ. 2018;27:725–30.
de Sousa VML, Carvalho L. Heterogeneity in lung cancer. Pathobiol J Immunopathol Mol Cell Biol. 2018;85:96–107.
Dai J, et al. Risk loci identification and polygenic risk score in prediction of lung cancer: a large-scale prospective cohort study in Chinese. Lancet Respir Med. 2019;7:881–91.
Hu Z, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792–6.
McKay JD, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017;49:1126–32.
Jin G, et al. Low-frequency coding variants at 6p21.33 and 20q11.21 are associated with lung cancer risk in Chinese populations. Am J Hum Genet. 2015;96:832–40.
Wu F, et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun. 2021;12:2540.
Marees AT, et al. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res. 2018;27: e1608.
Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Chang CC. Data management and summary statistics with PLINK. Methods Mol Biol Clifton NJ. 2020;2090:49–65.
Keich U, Noble WS. Controlling the FDR in imperfect matches to an incomplete database. J Am Stat Assoc. 2018;113:973–82.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Watanabe K, Umićević Mirkov M, de Leeuw CA, van den Heuvel MP, Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat Commun. 2019;10:3222.
Greener JG, Kandathil SM, Moffat L, Jones DT. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 2022;23:40–55.
Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015;349:255–60.
Uddin S, Khan A, Hossain ME, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak. 2019;19:281.
Thomas ZV, Wang Z, Zang C. BART Cancer: a web resource for transcriptional regulators in cancer genomes. NAR Cancer. 2021;3:zcab011.
Song S, Tang H, Quan W, Shang A, Ling C. Estradiol initiates the immune escape of non-small cell lung cancer cells via ERβ/SIRT1/FOXO3a/PD-L1 axis. Int Immunopharmacol. 2022;107: 108629.
Zhang Y, et al. MicroRNA-663a is downregulated in non-small cell lung cancer and inhibits proliferation and invasion by targeting JunD. BMC Cancer. 2016;16:315.
Sha Z, et al. Transcription factor CDX2 up-regulates proto-oncogenic miR-744 via a promoter activation mechanism in non-small-cell lung cancer. Ann Transl Med. 2021;9:1538.
Zeng Z, et al. Distinct expression and prognostic value of members of SMAD family in non-small cell lung cancer. Medicine. 2020;99: e19451.
Li J, Zhang S, Zhu L, Ma S. Role of transcription factor FOXA1 in non-small cell lung cancer. Mol Med Rep. 2018;17:509–21.
Kumar MS, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–55.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Feng, X. Integrative analysis of GWAS and transcriptomics data reveal key genes for non-small lung cancer. Med Oncol 40, 270 (2023). https://doi.org/10.1007/s12032-023-02139-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02139-x