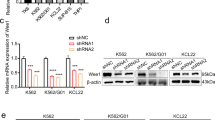
Abstract
The first-line drug Imatinib (IM) has achieved a curative effect in most chronic myeloid leukemia (CML) patients, but drug resistance remains a problem. More alternative therapeutic strategies need to explore. In recent years, targeting dysregulated DNA repair mechanisms provided promising options for cancer treatment. Here, we discovered the versatile Mediator of DNA Damage Checkpoint 1 (MDC1) interacted with γ-H2AX and 53BP1 in the early stage of the DNA damage response of cells. MDC1 overexpressed in CML cell lines and patients’ bone marrow mononuclear cells. By knocking down MDC1, non-homologous end-joining pathways were mainly inhibited, leading to an intense accumulation of unrepaired intracellular DNA damage and an apparent cell apoptosis promotion. Notably, targeting MDC1 further enhanced drug sensitivity in IM-resistant CML cells. Our work revealed that MDC1 is a prospective target for CML treatment through regulating DNA damage repair mechanism, and also an alternative option for IM resistance dilemma. This study extends the understanding of regulating dysfunctional DNA repair mechanisms for cancer treatment.






Similar content being viewed by others
References
de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp N, Groffen J, Stephenson JR. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–7.
Soverini S, Mancini M, Bavaro L, Cavo M, Martinelli G. Chronic myeloid leukemia: the paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol Cancer. 2018;17:49.
Bartram CR, de Klein A, Hagemeijer A, van Agthoven T, Geurts van Kessel A, Bootsma D, Grosveld G, Ferguson-Smith MA, Davies T, Stone M, et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306:277–80.
Clark RE. Tyrosine kinase inhibitor therapy discontinuation for patients with chronic myeloid leukaemia in clinical practice. Curr Hematol Malig Rep. 2019;14:507–14.
Braun TP, Eide CA, Druker BJ. Response and resistance to BCR-ABL1-targeted therapies. Cancer Cell. 2020;37:530–42.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Zhou H, Xu R. Leukemia stem cells: the root of chronic myeloid leukemia. Protein Cell. 2015;6:403–12.
Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45.
Popp HD, Kohl V, Naumann N, Flach J, Brendel S, Kleiner H, Weiss C, Seifarth W, Saussele S, Hofmann WK, Fabarius A. DNA damage and DNA damage response in chronic myeloid leukemia. Int J Mol Sci. 2020;21:1177.
Nowicki MO, Falinski R, Koptyra M, Slupianek A, Stoklosa T, Gloc E, Nieborowska-Skorska M, Blasiak J, Skorski T. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104:3746–53.
Skorski T. BCR/ABL regulates response to DNA damage: the role in resistance to genotoxic treatment and in genomic instability. Oncogene. 2002;21:8591–604.
Skorski T. BCR/ABL, DNA damage and DNA repair: implications for new treatment concepts. Leuk Lymphoma. 2008;49:610–4.
Kim TD, Türkmen S, Schwarz M, Koca G, Nogai H, Bommer C, Dörken B, Daniel P, le Coutre P. Impact of additional chromosomal aberrations and BCR-ABL kinase domain mutations on the response to nilotinib in Philadelphia chromosome-positive chronic myeloid leukemia. Haematologica. 2010;95:582–8.
Ozaki T, Nagase T, Ichimiya S, Seki N, Ohiri M, Nomura N, Takada N, Sakiyama S, Weber BL, Nakagawara A. NFBD1/KIAA0170 is a novel nuclear transcriptional transactivator with BRCT domain. DNA Cell Biol. 2000;19:475–85.
Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–6.
Bartkova J, Horejsí Z, Sehested M, Nesland JM, Rajpert-De Meyts E, Skakkebaek NE, Stucki M, Jackson S, Lukas J, Bartek J. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 2007;26:7414–22.
Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci USA. 2008;105:11200–5.
Nakanishi M, Ozaki T, Yamamoto H, Hanamoto T, Kikuchi H, Furuya K, Asaka M, Delia D, Nakagawara A. NFBD1/MDC1 associates with p53 and regulates its function at the crossroad between cell survival and death in response to DNA damage. J Biol Chem. 2007;282:22993–3004.
Dave JH, Vora HH, Ghosh NR, Trivedi TI. Mediator of DNA damage checkpoint protein 1 (MDC1) as a prognostic marker for patients with oral squamous cell carcinoma. J Oral Pathol Med. 2017;46:253–8.
Liu X, Qiu Z, Wang Z, Zuo W, Gong Z, Liu C, Zeng Q, Qian Y, Jiang L, Li Y, Bu Y, Hu G. NFBD1/MDC1 participates in the regulation of proliferation and apoptosis in human laryngeal squamous cell carcinoma. Clin Transl Oncol. 2018;20:534–41.
Liu X, Dong R, Jiang Z, Wei Y, Li Y, Wei L, Sun H, Li Y, Yang N, Yang Q, Liu Z, Kong B. MDC1 promotes ovarian cancer metastasis by inducing epithelial–mesenchymal transition. Tumour Biol. 2015;36:4261–9.
Yuan C, Bu Y, Wang C, Yi F, Yang Z, Huang X, Cheng L, Liu G, Wang Y, Song F. NFBD1/MDC1 is a protein of oncogenic potential in human cervical cancer. Mol Cell Biochem. 2012;359:333–46.
Zhu HL, Liu T, Meng WT, Jia YQ. Establishment of an Imatinib resistance cell line K562R and its resistant principia. Sichuan Da Xue Xue Bao Yi Xue. 2007;38:22–6.
Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4: e1000110.
Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–8.
Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–53.
Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem. 2018;293:10512–23.
Taleei R, Nikjoo H. The non-homologous end-joining (NHEJ) pathway for the repair of DNA double-strand breaks: I. A mathematical model. Radiat Res. 2013;179:530–9.
Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506.
O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60:547–60.
Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, Jasin M, Brunet E. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–42.
Rassool FV. DNA double strand breaks (DSB) and non-homologous end joining (NHEJ) pathways in human leukemia. Cancer Lett. 2003;193:1–9.
Zhu Y, Dai H, Wang Y, Liang Y, Feng W, Yuan Y. Targeting FEN1 suppresses the proliferation of chronic myeloid leukemia cells through regulating alternative end-joining pathways. DNA Cell Biol. 2021;40:1101–11.
Acknowledgements
We appreciate Dr.Yonghong Wang and Dr.Yalin Zhu for their assistance with experiments. Dr. Chen of the Radiology Department in the First Affiliated Hospital of Chongqing Medical University for the radiology support.
Funding
This work is supported by the National Natural Science Foundation of China (Grant No. 81703095), Chongqing Natural Science Foundation (Grant No. cstc2021jcyj-msxmX0214), Youth Top Science and Technology Talent Fund Project of The First Affiliated Hospital of Chongqing Medical University (Grant No. BJRC2020–04), and Innovation Support Program for Overseas Students of Chongqing (Grant No. cx2018142).
Author information
Authors and Affiliations
Contributions
YY and YL conceived and designed the experiments. YL and YQ completed the experiments and wrote the draft. YL, YQ, WF, and YY analyzed the data and results. YY, WF, and GJ supervised the project and revised the manuscript. The final version of the manuscript was read and approved by all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval and consent to participate
The research conformed to the standard stipulated by the Declaration of Helsinki and was performed with the approval of the Ethics Committee of Chongqing Medical University (ID:2022059).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12032_2022_1821_MOESM2_ESM.tif
Supplementary file2 (TIF 5166 KB)—Figure S1. (A) Relativity mRNA expression of MDC1 in various cancer cells, data from CCLE datasets. (B) Network analysis of known protein interaction partners of MDC1, from STRING website. (C) Left. Abnormal large K562/G01 cells (200×); Right. enlarged cell image. (D, E) Cell viability was detected by the CCK-8 assay. Data are expressed as the mean ± SD.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liang, Y., Qin, Y., Jiang, G. et al. Targeting MDC1 promotes apoptosis and sensitizes Imatinib resistance in CML cells by mainly disrupting non-homologous end-joining repair. Med Oncol 39, 226 (2022). https://doi.org/10.1007/s12032-022-01821-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01821-w