Abstract
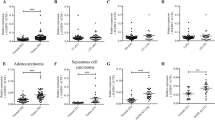
N-acetyltransferase 10 (NAT10) is a nucleolar acetyltransferase and has been reported to facilitate tumorigenesis in various cancers, but its role in NSCLC and how it is regulated remain to be assessed. The expression of NAT10 was explored in online databases and our collected clinical specimens. The relationship of NAT10 and clinical characteristics was evaluated using the online databases. Functional analyses were utilized to determine the effect of NAT10 on the proliferation and migration abilities. KEGG pathway analyses were conducted to investigate NAT10-related pathways in NSCLC. The influence of NAT10 on cell cycle was assessed by flow cytometry and cell synchronization assay. The association between c-myc and NAT10 promoter was determined by ChIP. Compared with normal tissue, NAT10 was significantly overexpressed in NSCLC. Upregulated NAT10 was associated with more advanced stage for lung adenocarcinoma and shorter overall survival and first progression time for lung cancer. NAT10 could promote proliferation and migration of NSCLC cells in vitro. c-myc positively regulated the expression of NAT10 as a transcription factor. KEGG pathway analyses indicated that NAT10 was significantly involved in cell cycle regulation, cytokine–cytokine receptor interaction and other pathways. The knockdown of NAT10-induced G1 arrest, which was possibly mediated by the downregulation of cyclin D1.Our findings suggested that c-myc-mediated upregulation of NAT10 promoted the proliferation and migration of NSCLC cells and NAT10 might be a marker for prognosis and a promising target for treatment in NSCLC.






Similar content being viewed by others
Data availability
The data used in this study are available from the corresponding author upon reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398(10299):535–54.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23(1):74–88.
Icard P, Fournel L, Wu Z, Alifano M, Lincet H. Interconnection between metabolism and cell cycle in cancer. Trends Biochem Sci. 2019;44(6):490–501.
Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175(7):1872-1886.e24.
Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28(5):773–85.
Shen Q, Zheng X, McNutt MA, Guang L, Sun Y, Wang J, et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res. 2009;315(10):1653–67.
Liu X, Cai S, Zhang C, Liu Z, Luo J, Xing B, et al. Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Res. 2018;46(18):9601–16.
Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17(3):349–66.
Cai S, Liu X, Zhang C, Xing B, Du X. Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem Biophys Res Commun. 2017;483(1):624–9.
Liu HY, Liu YY, Yang F, Zhang L, Zhang FL, Hu X, et al. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48(7):3638–56.
Zhang L, Li DQ. MORC2 regulates DNA damage response through a PARP1-dependent pathway. Nucleic Acids Res. 2019;47(16):8502–20.
Zheng J, Tan Y, Liu X, Zhang C, Su K, Jiang Y, et al. NAT10 regulates mitotic cell fate by acetylating Eg5 to control bipolar spindle assembly and chromosome segregation. Cell Death Differ. 2022;29(4):846–60.
Zhang Y, Jing Y, Wang Y, Tang J, Zhu X, Jin WL, et al. NAT10 promotes gastric cancer metastasis via N4-acetylated COL5A1. Signal Transduct Target Ther. 2021;6(1):173.
Zhang H, Hou W, Wang HL, Liu HJ, Jia XY, Zheng XZ, et al. GSK-3β-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin Cancer Res. 2014;20(17):4717–29.
Li Q, Liu X, Jin K, Lu M, Zhang C, Du X, et al. NAT10 is upregulated in hepatocellular carcinoma and enhances mutant p53 activity. BMC Cancer. 2017;17(1):605.
Tan Y, Zheng J, Liu X, Lu M, Zhang C, Xing B, et al. Loss of nucleolar localization of NAT10 promotes cell migration and invasion in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;499(4):1032–8.
Oh TI, Lee YM, Lim BO, Lim JH. Inhibition of NAT10 suppresses melanogenesis and melanoma growth by attenuating microphthalmia-associated transcription factor (MITF) expression. Int J Mol Sci. 2017. https://doi.org/10.3390/ijms18091924.
Wu J, Zhu H, Wu J, Chen W, Guan X. Inhibition of N-acetyltransferase 10 using remodelin attenuates doxorubicin resistance by reversing the epithelial-mesenchymal transition in breast cancer. Am J Transl Res. 2018;10(1):256–64.
Liang P, Hu R, Liu Z, Miao M, Jiang H, Li C. NAT10 upregulation indicates a poor prognosis in acute myeloid leukemia. Curr Probl Cancer. 2020;44(2):100491.
Zi J, Han Q, Gu S, McGrath M, Kane S, Song C, et al. Targeting NAT10 induces apoptosis associated with enhancing endoplasmic reticulum stress in acute myeloid leukemia cells. Front Oncol. 2020;10:598107.
Zhang X, Chen J, Jiang S, He S, Bai Y, Zhu L, et al. N-acetyltransferase 10 enhances doxorubicin resistance in human hepatocellular carcinoma cell lines by promoting the epithelial-to-mesenchymal transition. Oxid Med Cell Longev. 2019;2019:7561879.
Wu B, Chang N, Xi H, Xiong J, Zhou Y, Wu Y, et al. PHB2 promotes tumorigenesis via RACK1 in non-small cell lung cancer. Theranostics. 2021;11(7):3150–66.
Wang J, Wang C, Xu P, Li X, Lu Y, Jin D, et al. PRMT1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Theranostics. 2021;11(11):5387–403.
Zhou L, Xu Q, Huang L, Jin J, Zuo X, Zhang Q, et al. Low-dose carboplatin reprograms tumor immune microenvironment through STING signaling pathway and synergizes with PD-1 inhibitors in lung cancer. Cancer Lett. 2021;500:163–71.
Booy EP, McRae EK, Koul A, Lin F, McKenna SA. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol Cancer. 2017;16(1):109.
Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32(12):3683–8.
Apraiz A, Mitxelena J, Zubiaga A. Studying cell cycle-regulated gene expression by two complementary cell synchronization protocols. J Vis Exp. 2017. https://doi.org/10.3791/55745.
Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344(6183):527–32.
Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5(10):1024–39.
O’Connor MJ, Thakar T, Nicolae CM, Moldovan GL. PARP14 regulates cyclin D1 expression to promote cell-cycle progression. Oncogene. 2021;40(30):4872–83.
Cao Y, Yao M, Wu Y, Ma N, Liu H, Zhang B. N-Acetyltransferase 10 promotes micronuclei formation to activate the senescence-associated secretory phenotype machinery in colorectal cancer cells. Transl Oncol. 2020;13(8): 100783.
Tao W, Tian G, Xu S, Li J, Zhang Z, Li J. NAT10 as a potential prognostic biomarker and therapeutic target for HNSCC. Cancer Cell Int. 2021;21(1):413.
Wu Y, Cao Y, Liu H, Yao M, Ma N, Zhang B. Remodelin, an inhibitor of NAT10, could suppress hypoxia-induced or constitutional expression of HIFs in cells. Mol Cell Biochem. 2020;472(1–2):19–31.
Suski JM, Braun M, Strmiska V, Sicinski P. Targeting cell-cycle machinery in cancer. Cancer Cell. 2021;39(6):759–78.
Tchakarska G, Sola B. The double dealing of cyclin D1. Cell Cycle. 2020;19(2):163–78.
Liu H, Ling Y, Gong Y, Sun Y, Hou L, Zhang B. DNA damage induces N-acetyltransferase NAT10 gene expression through transcriptional activation. Mol Cell Biochem. 2007;300(1–2):249–58.
Jirawatnotai S, Hu Y, Livingston DM, Sicinski P. Proteomic identification of a direct role for cyclin d1 in DNA damage repair. Cancer Res. 2012;72(17):4289–93.
Dong P, Maddali MV, Srimani JK, Thélot F, Nevins JR, Mathey-Prevot B, et al. Division of labour between Myc and G1 cyclins in cell cycle commitment and pace control. Nat Commun. 2014;5:4750.
Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97(5):2229–34.
Beier R, Bürgin A, Kiermaier A, Fero M, Karsunky H, Saffrich R, et al. Induction of cyclin E-cdk2 kinase activity, E2F-dependent transcription and cell growth by Myc are genetically separable events. Embo J. 2000;19(21):5813–23.
Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849(5):506–16.
Curti L, Campaner S. MYC-induced replicative stress: a double-edged sword for cancer development and treatment. Int J Mol Sci. 2021. https://doi.org/10.3390/ijms22126168.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81770082) and the Natural Science Foundation of Jiangsu Province (BE2019719).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by ZW, YH, WL and JL. XL and SZ contributed to the interpretation of data. The first draft of the manuscript was written by ZW. HL and YS supervised the whole study. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Z., Huang, Y., Lu, W. et al. c-myc-mediated upregulation of NAT10 facilitates tumor development via cell cycle regulation in non-small cell lung cancer. Med Oncol 39, 140 (2022). https://doi.org/10.1007/s12032-022-01736-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01736-6