Abstract
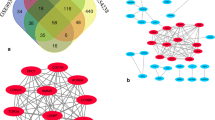
Mechanisms underlying the regulation of gene expression in cancer have been surveyed for decades to find novel prognostic factors and new targets for molecular targeted therapies in cancer. Because most cases of liver cancer are associated with liver cirrhosis, we aimed to analyze the gene expression signatures and the gene regulatory mechanism in hepatocellular carcinoma (HCC) on a cirrhotic background using high-throughput data analysis. In the present study, three valid array-based datasets containing HCC and liver cirrhosis samples were obtained to identify common differentially expressed genes (DEGs). Moreover, a comprehensive data analysis was conducted based on RNA-Seq data and using Kaplan–Meier curve analysis to find molecular signatures that reduce patients' survival rate. Furthermore, we proposed a gene regulatory network (GRN) to explore the possible regulatory mechanism of these molecular signatures by transcription factors in HCC progression from cirrhosis. Besides, we analyzed protein–protein interactions, gene ontology (GO), and pathway enrichment to elucidate the cellular and molecular function of the GRN elements in HCC. In this way, we found a list of 231 molecular signatures in HCC derived from cirrhosis. We also found the importance of TCF4, RUNX1, HINFP, KDM2B, MAF, JUN, NR5A2, NFYA, and AR as key differentially expressed transcription factors (DETFs) in the progression of HCC from cirrhosis. In conclusion, the identified molecular signatures and their transcription factors propose candidate prognostic markers and possible molecular targets in the progression of HCC.






Similar content being viewed by others
Data availability
All data will be made available on request.
References
Siegel RL, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
Sahu SK, et al. Rupture of hepatocellular carcinoma: a review of literature. J Clin Exp Hepatol. 2019;9(2):245–56.
Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Rees CJ, Koo S. Artificial intelligence—upping the game in gastrointestinal endoscopy? Nat Rev Gastroenterol Hepatol. 2019;16(10):584–5.
Janevska D, Chaloska-Ivanova V, Janevski V. Hepatocellular carcinoma: risk factors, diagnosis and treatment. Open Access Maced J Med Sci. 2015;3(4):732.
Fan F, et al. Withdrawal of immunosuppressive therapy in allogeneic bone marrow transplantation reactivates chronic viral hepatitis C. Bone Marrow Transplant. 1991;8(5):417–20.
Rocio G, et al. Insulin-like growth factor-1 deficiency and cirrhosis establishment. J CliN Med Res. 2017;9(4):233.
Oliveira CP, et al. Nutrition and physical activity in nonalcoholic fatty liver disease. J Diabetes Res. 2016. https://doi.org/10.1155/2016/4597246.
Ramakrishna G, et al. From cirrhosis to hepatocellular carcinoma: new molecular insights on inflammation and cellular senescence. Liver Cancer. 2013;2(3–4):367–83.
Chen C, Wang G. Mechanisms of hepatocellular carcinoma and challenges and opportunities for molecular targeted therapy. World J Hepatol. 2015;7(15):1964.
Balmain A. Cancer genetics: from Boveri and Mendel to microarrays. Nat Rev Cancer. 2001;1(1):77–82.
de Sousa Abreu R, et al. Global signatures of protein and mRNA expression levels. Mol BioSyst. 2009;5(12):1512–26.
Sung W-K, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44(7):765–9.
Kojima K, et al. Transcriptome profiling of archived sectioned formalin-fixed paraffin-embedded (AS-FFPE) tissue for disease classification. PloS One. 2014;9(1):e86961.
Moeini A, et al. An immune gene expression signature associated with development of human hepatocellular carcinoma identifies mice that respond to chemopreventive agents. Gastroenterology. 2019;157(5):1383-1397.e11.
Lamb JR, et al. Predictive genes in adjacent normal tissue are preferentially altered by sCNV during tumorigenesis in liver cancer and may rate limiting. PloS One. 2011;6(7):e20090.
Smyth GK. Limma: linear models for microarray data. In: Bioinformatics and computational biology solutions using R and Bioconductor. Springer; 2005. p. 397–420.
Gong Q, Fang L. Asymptotic properties of mean survival estimate based on the Kaplan-Meier curve with an extrapolated tail. Pharm Stat. 2012;11(2):135–40.
Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5(12):181006.
Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–44.
Scardoni G, et al. Biological network analysis with CentiScaPe: centralities and experimental dataset integration. F1000Res. 2014;3:139.
Zhou, Z., et al., 2020 Transcriptome analysis of the cytokinin response in Medicago truncatula. Journal of Plant Biology, 1–14.
Eskandari E, Motalebzadeh J. Transcriptomics-based screening of molecular signatures associated with patients overall survival and their key regulators in subtypes of breast cancer. Cancer Genet. 2019;239:62–74.
Szklarczyk D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13.
Lu Y, et al. Analysis of long non-coding RNA expression profiles identifies functional lncRNAs associated with the progression of acute coronary syndromes. Exp Ther Med. 2018;15(2):1376–84.
Long T, et al. Identification of differentially expressed genes and enriched pathways in lung cancer using bioinformatics analysis. Mol Med Rep. 2019;19(3):2029–40.
Gene Ontology Consortium. The Gene ontology (GO) database and informatics resource. Nucleic AcidS Res. 2004;32(suppl_1):D258–61.
Abdelzaher AF, et al. Transcriptional network growing models using motif-based preferential attachment. Front Bioeng Biotechnol. 2015;3:157.
Pinter M, et al. Cancer and liver cirrhosis: implications on prognosis and management. ESMO Open. 2016;1(2):e000042.
Bosveld F, Wang Z, Bellaïche Y. Tricellular junctions: a hot corner of epithelial biology. Curr Opin Cell Biol. 2018;54:80–8.
Wang E, Lenferink A, O’Connor-McCourt M. Cancer systems biology: exploring cancer-associated genes on cellular networks. Cell Mol Life Sci. 2007. https://doi.org/10.1007/s00018-007-7054-6.
Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80.
Sánchez-Tilló E, et al. β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci. 2011;108(48):19204–9.
Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129(15):2070–82.
Hong D, et al. RUNX1-dependent mechanisms in biological control and dysregulation in cancer. J Cell Physiol. 2019;234(6):8597–609.
Yan M, et al. The critical role of histone lysine demethylase KDM2B in cancer. Am J Transl Res. 2018;10(8):2222.
Bezzecchi E, et al. NF-Y overexpression in liver Hepatocellular carcinoma (HCC). Int J Mol Sci. 2020;21(23):9157.
Eskandari E, Mahjoubi F, Motalebzadeh J. An integrated study on TFs and miRNAs in colorectal cancer metastasis and evaluation of three co-regulated candidate genes as prognostic markers. Gene. 2018;679:150–9.
Yang Q, et al. NR5A2 promotes cell growth and resistance to temozolomide through regulating notch signal pathway in glioma. Onco Targets Ther. 2020;13:10231.
Kanda T, Jiang X, Yokosuka O. Androgen receptor signaling in hepatocellular carcinoma and pancreatic cancers. World J Gastroenterol. 2014;20(28):9229.
Snider J, et al. Fundamentals of protein interaction network mapping. Mol Syst Biol. 2015;11(12):848.
Cai W, et al. PMP22 regulates self-renewal and chemoresistance of gastric cancer cells. Mol Cancer Ther. 2017;16(6):1187–98.
Bacon CM, Du M-Q, Dogan A. Mucosa-associated lymphoid tissue (MALT) lymphoma: a practical guide for pathologists. J Clin Pathol. 2007;60(4):361–72.
Cui H, et al. Gamma linolenic acid regulates PHD2 mediated hypoxia and mitochondrial apoptosis in DEN induced hepatocellular carcinoma. Drug Des Dev Ther. 2018;12:4241.
Santoro N, Caprio S, Feldstein AE. Oxidized metabolites of linoleic acid as biomarkers of liver injury in nonalcoholic steatohepatitis. Clin Lipidol. 2013;8(4):411–8.
Acknowledgements
This research was conducted using the authors’ personal money.
Author information
Authors and Affiliations
Contributions
JM participated in the conceptualization, investigation, formal analysis, writing, reviewing, and editing of the original draft, and supervision. EE participated in the investigation, formal analysis, and writing of the original draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Motalebzadeh, J., Eskandari, E. Transcription factors linked to the molecular signatures in the development of hepatocellular carcinoma on a cirrhotic background. Med Oncol 38, 121 (2021). https://doi.org/10.1007/s12032-021-01567-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-021-01567-x