Abstract
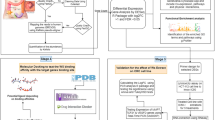
Ovarian cancer is the first leading cause of mortality in gynecological malignancies. To identify key genes and microRNAs in ovarian cancer, mRNA microarray dataset GSE36668, GSE18520, GSE14407 and microRNA dataset GSE47841 were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) and microRNAs (DEMs) were obtained using GEO2R. Functional and pathway enrichment analysis were performed for DEGs using DAVID database. Protein–protein interaction (PPI) network was established by STRING and visualized by Cytoscape. Following, overall survival (OS) analysis of hub genes was performed by the Kaplan–Meier plotter online tool. Module analysis of the PPI network was performed using MCODE. Moreover, miRecords was applied to predict the targets of the DEMs. A total of 345 DEGs were obtained, which were mainly enriched in the terms related to cell cycle, mitosis, and ovulation cycle process. A PPI network was constructed, consisting of 141 nodes and 296 edges. Sixteen genes had high degrees in the network. High expression of four genes of the 16 genes was associated with worse OS of patients with ovarian cancer, including CCNB1, CENPF, KIF11, and ZWINT. A significant module was detected from the PPI network. The enriched functions and pathways included cell cycle, nuclear division, and oocyte meiosis. Additionally, a total of 36 DEMs were identified. The expression of KIF11 was negatively correlated with that of has-miR-424 and has-miR-381, and it was also the potential target of two microRNAs. In conclusion, these results identified key genes, which could provide potential targets for ovarian cancer diagnosis and treatment.



Similar content being viewed by others
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262.
Wright JD, Shah M, Mathew L, Burke WM, Culhane J, Goldman N, et al. Fertility preservation in young women with epithelial ovarian cancer. Cancer. 2009;115(18):4118–26. doi:10.1002/cncr.24461.
Urban N, Drescher C. Potential and limitations in early diagnosis of ovarian cancer. Adv Exp Med Biol. 2008;622:3–14. doi:10.1007/978-0-387-68969-2_1.
Goff BA, Mandel L, Muntz HG, Melancon CH. Ovarian carcinoma diagnosis. Cancer. 2000;89(10):2068–75.
Urban N, Hawley S, Janes H, Karlan BY, Berg CD, Drescher CW, et al. Identifying post-menopausal women at elevated risk for epithelial ovarian cancer. Gynecol Oncol. 2015;139(2):253–60. doi:10.1016/j.ygyno.2015.08.024.
Felder M, Kapur A, Gonzalez-Bosquet J, Horibata S, Heintz J, Albrecht R, et al. MUC16 (CA125): tumor biomarker to cancer therapy, a work in progress. Mol Cancer. 2014;13:129. doi:10.1186/1476-4598-13-129.
Vathipadiekal V, Wang V, Wei W, Waldron L, Drapkin R, Gillette M, et al. Creation of a human secretome: a novel composite library of human secreted proteins—validation using ovarian cancer gene expression data and a virtual secretome array. Clin Cancer Res. 2015;21(21):4960–9. doi:10.1158/1078-0432.CCR-14-3173.
Wei SU, Li H, Zhang B. The diagnostic value of serum HE4 and CA-125 and ROMA index in ovarian cancer. Biomed Rep. 2016;5(1):41–4. doi:10.3892/br.2016.682.
Bowen NJ, Walker LD, Matyunina LV, Logani S, Totten KA, Benigno BB, et al. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med Genom. 2009;2:71. doi:10.1186/1755-8794-2-71.
Elgaaen BV, Olstad OK, Sandvik L, Odegaard E, Sauer T, Staff AC, et al. ZNF385B and VEGFA are strongly differentially expressed in serous ovarian carcinomas and correlate with survival. PLoS One. 2012;7(9):e46317. doi:10.1371/journal.pone.0046317.
Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong KK, et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell. 2009;16(6):521–32. doi:10.1016/j.ccr.2009.10.018.
Elgaaen BV, Olstad OK, Haug KB, Brusletto B, Sandvik L, Staff AC, et al. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer. 2014;14:80. doi:10.1186/1471-2407-14-80.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41(Database issue):D991–5. doi:10.1093/nar/gks1193.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. doi:10.1093/nar/gku1003.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genom Res. 2003;13(11):2498–504. doi:10.1101/gr.1239303.
Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2.
Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Database issue):D105–10. doi:10.1093/nar/gkn851.
Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19(2):197–208. doi:10.1530/ERC-11-0329.
Wan J, Shi F, Xu Z, Zhao M. Knockdown of eIF4E suppresses cell proliferation, invasion and enhances cisplatin cytotoxicity in human ovarian cancer cells. Int J Oncol. 2015;47(6):2217–25. doi:10.3892/ijo.2015.3201.
Fang Y, Yu H, Liang X, Xu J, Cai X. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther. 2014;15(9):1268–79. doi:10.4161/cbt.29691.
Zhou L, Li J, Zhao YP, Cui QC, Zhou WX, Guo JC, et al. The prognostic value of Cyclin B1 in pancreatic cancer. Med Oncol. 2014;31(9):107. doi:10.1007/s12032-014-0107-4.
Avery-Kiejda KA, Bowden NA, Croft AJ, Scurr LL, Kairupan CF, Ashton KA, et al. P53 in human melanoma fails to regulate target genes associated with apoptosis and the cell cycle and may contribute to proliferation. BMC Cancer. 2011;11:203. doi:10.1186/1471-2407-11-203.
Kreis NN, Friemel A, Zimmer B, Roth S, Rieger MA, Rolle U, et al. Mitotic p21Cip1/CDKN1A is regulated by cyclin-dependent kinase 1 phosphorylation. Oncotarget. 2016. doi:10.18632/oncotarget.10330.
Tu Y, Kim E, Gao Y, Rankin GO, Li B, Chen YC. Theaflavin-3, 3′-digallate induces apoptosis and G2 cell cycle arrest through the Akt/MDM2/p53 pathway in cisplatin-resistant ovarian cancer A2780/CP70 cells. Int J Oncol. 2016;48(6):2657–65. doi:10.3892/ijo.2016.3472.
Zhou J, Zhao M, Tang Y, Wang J, Wei C, Gu F, et al. The milk-derived fusion peptide, ACFP, suppresses the growth of primary human ovarian cancer cells by regulating apoptotic gene expression and signaling pathways. BMC Cancer. 2016;16:246. doi:10.1186/s12885-016-2281-6.
Volkov VA, Grissom PM, Arzhanik VK, Zaytsev AV, Renganathan K, McClure-Begley T, et al. Centromere protein F includes two sites that couple efficiently to depolymerizing microtubules. J Cell Biol. 2015;209(6):813–28. doi:10.1083/jcb.201408083.
Brendle A, Brandt A, Johansson R, Enquist K, Hallmans G, Hemminki K, et al. Single nucleotide polymorphisms in chromosomal instability genes and risk and clinical outcome of breast cancer: a Swedish prospective case–control study. Eur J Cancer. 2009;45(3):435–42. doi:10.1016/j.ejca.2008.10.001.
Dai Y, Liu L, Zeng T, Zhu YH, Li J, Chen L, et al. Characterization of the oncogenic function of centromere protein F in hepatocellular carcinoma. Biochem Biophys Res Commun. 2013;436(4):711–8. doi:10.1016/j.bbrc.2013.06.021.
Zhuo YJ, Xi M, Wan YP, Hua W, Liu YL, Wan S, et al. Enhanced expression of centromere protein F predicts clinical progression and prognosis in patients with prostate cancer. Int J Mol Med. 2015;35(4):966–72. doi:10.3892/ijmm.2015.2086.
Martens-de Kemp SR, Nagel R, Stigter-van Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ, et al. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clin Cancer Res. 2013;19(8):1994–2003. doi:10.1158/1078-0432.CCR-12-2539.
Venere M, Horbinski C, Crish JF, Jin X, Vasanji A, Major J, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med. 2015;7(304):304ra143. doi:10.1126/scitranslmed.aac6762.
Mirzaa GM, Enyedi L, Parsons G, Collins S, Medne L, Adams C, et al. Congenital microcephaly and chorioretinopathy due to de novo heterozygous KIF11 mutations: five novel mutations and review of the literature. Am J Med Genetics Part A. 2014;164A(11):2879–86. doi:10.1002/ajmg.a.36707.
Seo DW, You SY, Chung WJ, Cho DH, Kim JS, Oh JS. Zwint-1 is required for spindle assembly checkpoint function and kinetochore-microtubule attachment during oocyte meiosis. Sci Rep. 2015;5:15431. doi:10.1038/srep15431.
Endo H, Ikeda K, Urano T, Horie-Inoue K, Inoue S. Terf/TRIM17 stimulates degradation of kinetochore protein ZWINT and regulates cell proliferation. J Biochem. 2012;151(2):139–44. doi:10.1093/jb/mvr128.
Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi:10.1016/j.addr.2015.05.001.
Zuberi M, Mir R, Das J, Ahmad I, Javid J, Yadav P, et al. Erratum to: Expression of serum miR-200a, miR-200b and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol. 2015;17(10):840. doi:10.1007/s12094-015-1355-2.
Meng X, Muller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7(13):16923–35. doi:10.18632/oncotarget.7850.
Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016;7:11406. doi:10.1038/ncomms11406.
Xia B, Li H, Yang S, Liu T, Lou G. MiR-381 inhibits epithelial ovarian cancer malignancy via YY1 suppression. Tumour Biol. 2016. doi:10.1007/s13277-016-4805-8.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Xu, Z., Zhou, Y., Cao, Y. et al. Identification of candidate biomarkers and analysis of prognostic values in ovarian cancer by integrated bioinformatics analysis. Med Oncol 33, 130 (2016). https://doi.org/10.1007/s12032-016-0840-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-016-0840-y