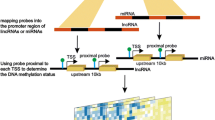
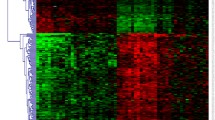
Abstract
Hippocampal sclerosis (HS) is one of the most prevalent pathological types of temporal lobe epilepsy (TLE), and it significantly affects patient prognoses. The methylation of DNA plays an important role in the development of epilepsy. However, few studies have focused on HS subtypes to determine DNA methylation profiles in TLE. This study aimed to determine the pathogenesis of TLE from an epigenetic perspective in patients with TLE-HS type I (TLE-HSTI) and TLE without HS (TLE-nHS) using whole-genome bisulfite sequencing (WGBS). We defined 1171 hypermethylated and 2537 hypomethylated regions and found 632 differentially methylated genes (DMG) in the promoter region that were primarily involved in the regulation of various aspects of epilepsy development. Twelve DMG overlapped with differentially expressed genes (DEG) in the promoter region, and RT-qPCR findings revealed significant overexpression of the SBNO2, CBX3, RASAL3, and TMBIM4 genes in TLE-HSTI. We present the first systematic analysis of methylation profiles of TLE-HSTI and TLE-nHS from an epigenetic perspective using WGBS. Overall, our preliminary data highlight the underlying mechanism of TLE-HSTI, providing a new perspective for guiding treatment of TLE.







Similar content being viewed by others
Availability of Data and Material
The raw data are publicly available in the GEO, under accession number GSE140658.
References
Asadi-Pooya AA, Nei M, Sharan A, Sperling MR (2016) Auras in patients with temporal lobe epilepsy and mesial temporal sclerosis. J Neurol Sci 364:24–26. https://doi.org/10.1016/j.jns.2016.03.006
Barker-Haliski M, White HS (2015) Glutamatergic Mechanisms Associated with Seizures and Epilepsy. Cold Spring Harb Perspect Med 5:a022863. https://doi.org/10.1101/cshperspect.a022863
Blümcke I et al (2007) A new clinico-pathological classification system for mesial temporal sclerosis. Acta Neuropathol 113:235–244. https://doi.org/10.1007/s00401-006-0187-0
Blumcke I et al (2013) International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: a Task Force report from the ILAE Commission on Diagnostic Methods. Epilepsia 54:1315–1329. https://doi.org/10.1111/epi.12220
Calderon-Garciduenas AL et al (2018) New clinicopathological associations and histoprognostic markers in ILAE types of hippocampal sclerosis. Brain Pathol 28:644–655. https://doi.org/10.1111/bpa.12596
Feng H, Conneely KN, Wu H (2014) A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res 42:e69. https://doi.org/10.1093/nar/gku154
Feng Y-CA et al (2019) Ultra-Rare Genetic Variation in the Epilepsies: A Whole-Exome Sequencing Study of 17,606 Individuals. Am J Human Genet 105:267–282. https://doi.org/10.1016/j.ajhg.2019.05.020
Gales JM, Jehi L, Nowacki A, Prayson RA (2017) The role of histopathologic subtype in the setting of hippocampal sclerosis-associated mesial temporal lobe epilepsy. Hum Pathol 63:79–88. https://doi.org/10.1016/j.humpath.2017.02.013
Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128:635–638. https://doi.org/10.1016/j.cell.2007.02.006
Griffin NG et al (2016) Differential gene expression in dentate granule cells in mesial temporal lobe epilepsy with and without hippocampal sclerosis. Epilepsia 57:376–385. https://doi.org/10.1111/epi.13305
Guelfi S et al (2019) Transcriptomic and genetic analyses reveal potential causal drivers for intractable partial epilepsy. Brain 142:1616–1630. https://doi.org/10.1093/brain/awz074
Hama N et al (2018) Epigenetic landscape influences the liver cancer genome architecture. Nat Commun 9:1643. https://doi.org/10.1038/s41467-018-03999-y
Kanehisa M et al (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480-484. https://doi.org/10.1093/nar/gkm882
Kasperaviciute D et al (2010) Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain 133:2136–2147. https://doi.org/10.1093/brain/awq130
Kasperavičiūtė D et al (2013) Epilepsy, hippocampal sclerosis and febrile seizures linked by common genetic variation around SCN1A. Brain 136:3140–3150. https://doi.org/10.1093/brain/awt233
Kwan P et al (2010) Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia 51:1069–1077. https://doi.org/10.1111/j.1528-1167.2009.02397.x
Kwan P, Brodie MJ (2000) Early identification of refractory epilepsy. N Engl J Med 342:314–319. https://doi.org/10.1056/NEJM200002033420503
Lai Y, Hu X, Chen G, Wang X, Zhu B (2016) Down-regulation of adenylate kinase 5 in temporal lobe epilepsy patients and rat model. J Neurol Sci 366:20–26. https://doi.org/10.1016/j.jns.2016.04.037
Li J et al (2019) Guide Positioning Sequencing identifies aberrant DNA methylation patterns that alter cell identity and tumor-immune surveillance networks. Genome Res 29:270–280. https://doi.org/10.1101/gr.240606.118
Lister R et al (2013) Global epigenomic reconfiguration during mammalian brain development. Sci 341:1237905. https://doi.org/10.1126/science.1237905
Lu JQ, Steve TA, Wheatley M, Gross DW (2017) Immune Cell Infiltrates in Hippocampal Sclerosis: Correlation With Neuronal Loss. J Neuropathol Exp Neurol 76:206–215. https://doi.org/10.1093/jnen/nlx001
Malmgren K, Thom M (2012) Hippocampal sclerosis--origins and imaging. Epilepsia 53(Suppl 4):19–33. https://doi.org/10.1111/j.1528-1167.2012.03610.x
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinform 21:3787–3793. https://doi.org/10.1093/bioinformatics/bti430
Mathon B et al (2017) Predictive factors of long-term outcomes of surgery for mesial temporal lobe epilepsy associated with hippocampal sclerosis. Epilepsia 58:1473–1485. https://doi.org/10.1111/epi.13831
Miller-Delaney SF et al (2015) Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain 138:616–631. https://doi.org/10.1093/brain/awu373
Mota MVB et al (2019) ATP Synthase Subunit Beta Immunostaining is Reduced in the Sclerotic Hippocampus of Epilepsy Patients. Cell Mol Neurobiol 39:149–160. https://doi.org/10.1007/s10571-018-0641-2
Park Y, Wu H (2016) Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 32:1446–1453. https://doi.org/10.1093/bioinformatics/btw026
Schenk GJ et al (2010) Over-expression of the DCLK gene transcript CARP decreases CA3/CA1 network excitability. Brain Res 1352:21–34. https://doi.org/10.1016/j.brainres.2010.07.068
Tang C et al (2019) The MicroRNA Expression Profiles of Human Temporal Lobe Epilepsy in HS ILAE Type 1. Cell Mol Neurobiol 39:461–470. https://doi.org/10.1007/s10571-019-00662-y
Thom M (2014) Review: Hippocampal sclerosis in epilepsy: a neuropathology review. Neuropathol Appl Neurobiol 40:520–543. https://doi.org/10.1111/nan.12150
Thurman DJ et al (2011) Standards for epidemiologic studies and surveillance of epilepsy. Epilepsia 52(Suppl 7):2–26. https://doi.org/10.1111/j.1528-1167.2011.03121.x
Wang C, Hong Z, Chen Y (2015) Involvement of p38 MAPK in the Drug Resistance of Refractory Epilepsy Through the Regulation Multidrug Resistance-Associated Protein 1. Neurochem Res 40:1546–1553. https://doi.org/10.1007/s11064-015-1617-y
Weinberg DN et al (2019) The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nat 573:281–286. https://doi.org/10.1038/s41586-019-1534-3
Wu DM, Zhang YT, Lu J, Zheng YL (2018) Effects of microRNA-129 and its target gene c-Fos on proliferation and apoptosis of hippocampal neurons in rats with epilepsy via the MAPK signaling pathway. J Cell Physiol 233:6632–6643. https://doi.org/10.1002/jcp.26297
Wu H et al (2015) Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res 43:e141. https://doi.org/10.1093/nar/gkv715
Yang N et al (2019) Dexamethasone ameliorates the damage of hippocampal filamentous actin cytoskeleton but is not sufficient to cease epileptogenesis in pilocarpine induced epileptic mice. Epilepsy Res 154:26–33. https://doi.org/10.1016/j.eplepsyres.2019.04.009
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Yu S et al (2014) Long-term outcome of epilepsy surgery: a retrospective study in a population of 379 cases. Epilepsy Res 108:555–564. https://doi.org/10.1016/j.eplepsyres.2013.12.004
Zhang M, Zhou X, Jiang W, Li M, Zhou R, Zhou S (2020) AJAP1 affects behavioral changes and GABABR1 level in epileptic mice. Biochem Biophys Res Commun 524:1057–1063. https://doi.org/10.1016/j.bbrc.2020.02.036
Zhang YF, Xiong TQ, Tan BH, Song Y, Li SL, Yang LB, Li YC (2014) Pilocarpine-induced epilepsy is associated with actin cytoskeleton reorganization in the mossy fiber-CA3 synapses. Epilepsy Res 108:379–389. https://doi.org/10.1016/j.eplepsyres.2014.01.016
Acknowledgments
We are grateful to all the patients for their participation in the investigation, and the staff for excellent technical assistance and sample collection.
Funding
This work was supported by the National Nature Science Foundation of China (no. 81601130, U20A20383 and 81571646), Natural Science Foundation of Heilongjiang Province (no. LH2019H082), Certificate of China Postdoctoral Science Foundation Grant (no. 2018M631951), Financial Assistance under Heilongjiang Postdoctoral Fund (no. LBH-Z17144), and Innovation Fund of the First Affiliated Hospital of Harbin Medical University (no. 2020L06).
Author information
Authors and Affiliations
Contributions
Meng Na, Zhiguo Lin, and Hong Shen conceived and designed the study; Chongyang Tang and Zhenfeng Jiang analyzed the data; Miaomiao Jiang, Shi Yan, and Haiyang Wang carried out immunohistochemical experiments; Wang Zhang created the figures and wrote the manuscript; Alicia Y. Hou revised the language of the manuscript; Binchao Liu, Xian Han, and Yifei Gu conducted the assays and contributed to data acquisition. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors have no conflicts of interest to declare.
Ethics Approval and Consent to Participate
All subjects in this study provided their written informed consent. The study protocol was approved by the Ethics Committee of the First Clinical College of Harbin Medical University.
Consent for Publication
Written informed consent for publication was obtained from all participants.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12031_2020_1780_MOESM1_ESM.tif
Supplementary file1 Supplementary Figure 1 Classification of methylated cytosines. Three samples each from patients with TLE-HSTI (A, B, and C), and TLE-nHS (D, E, and F). Colors represent methylated C sites under different contexts, and the area of each part represents the proportion of methylated C sites under the corresponding context. C, cytosine (TIF 490 KB)
12031_2020_1780_MOESM2_ESM.tif
Supplementary file2 Supplementary Figure 2 Heat map of DNA methylation profiles in sequence context. CGI, CpG island (TIF 1373 KB)
12031_2020_1780_MOESM3_ESM.tif
Supplementary file3 Supplementary Figure 3 Distribution of DMR among genomic regions. CGI, CpG island; DMR, differentially expressed regions (TIF 1151 KB)
12031_2020_1780_MOESM4_ESM.tif
Supplementary file4 Supplementary Figure 4 Gene Ontology classification and KEGG pathway enrichment of DMG based on CG (A, B) and in promoter region (C, D). CG, cytosine-guanine DMG, differentially methylated genes; KEGG, Kyoto Encyclopedia of Genes and Genomes (TIF 1229 KB)
Rights and permissions
About this article
Cite this article
Zhang, W., Wang, H., Liu, B. et al. Differential DNA Methylation Profiles in Patients with Temporal Lobe Epilepsy and Hippocampal Sclerosis ILAE Type I. J Mol Neurosci 71, 1951–1966 (2021). https://doi.org/10.1007/s12031-020-01780-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-020-01780-9