Abstract
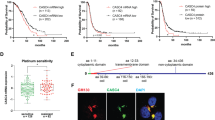
Gasdermin proteins (GSDMs) form pores in cell membranes upon various stimuli, leading to the release of certain proinflammatory molecules such as IL-1β and IL-18, and this ultimately results in pyroptotic cell death. NINJ1 (Ninjurin 1) has recently been identified as a cell membrane protein responsible for the final complete plasma membrane rupture following lytic cell death mechanisms including pyroptosis, causing the release of relatively larger molecules such as HMGB1 and LDH. In this study, we reported the presence of higher GSDMD and lower GSDME protein levels in ovarian tumors compared to surrounding non-malignant stroma in the tumor microenvironment. GSDME protein levels are also lower in the tumors of the omentum compared to adjacent stromal cells. We found that NINJ1 expression decreases from early to late stage in serous ovarian cancer, and the percentage of NINJ1 copy number loss events is the highest in ovarian cancer among other cancers. Moreover, we showed that low expression of NINJ1 is associated with shorter overall survival of patients with ovarian cancer. In support of the findings showing that low NINJ1 expression contributes to worse prognosis in this most lethal gynecological malignancy, NINJ1 expression was found to be lower in cisplatin-resistant ovarian cancer cells compared to cisplatin-sensitive counterparts in vitro. We suggest that the members of gasdermin family might have distinct functions in serous ovarian cancer, and low levels of NINJ1 might contribute, at least in part, to the progression and poorer prognosis of ovarian cancer. A complete picture of how pyroptosis and subsequent plasma membrane rupture are involved in ovarian cancer will be of high importance in order to identify actionable therapeutic vulnerabilities within this newly identified group of proteins.








Similar content being viewed by others
Data availability
No new data is generated in the current study.
Code availability
R code is available as a supplementary file.
References
Siegel RL, Miller KD. Jemal A 2016 Cancer statistics. CA Cancer J Clin. 2016;66(1):7–30. https://doi.org/10.3322/caac.21332.
Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian Cancer Nat Rev Dis Primers. 2016;25(2):16061. https://doi.org/10.1038/nrdp.2016.61.
Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A. Siegel RL 2018 Ovarian cancer statistics. CA Cancer J Clin. 2018;68(4):284–96. https://doi.org/10.3322/caac.21456.
Berkel C, Cacan E. GAB2 and GAB3 are expressed in a tumor stage-, grade- and histotype-dependent manner and are associated with shorter progression-free survival in ovarian cancer. J Cell Commun Signal. 2021;15(1):57–70. https://doi.org/10.1007/s12079-020-00582-3.
Gadducci A, Guarneri V, Peccatori FA, Ronzino G, Scandurra G, Zamagni C, Zola P, Salutari V. Current strategies for the targeted treatment of high-grade serous epithelial ovarian cancer and relevance of BRCA mutational status. J Ovarian Res. 2019;12(1):9. https://doi.org/10.1186/s13048-019-0484-6.
Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526(7575):666–71. https://doi.org/10.1038/nature15541.
Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–5. https://doi.org/10.1038/nature15514.
He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015;25(12):1285–98. https://doi.org/10.1038/cr.2015.139.
Broz P, Pelegrín P, Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 2020;20(3):143–57. https://doi.org/10.1038/s41577-019-0228-2.
Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535(7610):111–6. https://doi.org/10.1038/nature18590.
Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, Dueber EC. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci USA. 2016;113(28):7858–63. https://doi.org/10.1073/pnas.1607769113.
Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535(7610):153–8. https://doi.org/10.1038/nature18629.
Sborgi L, Rühl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, Farady CJ, Müller DJ, Broz P, Hiller S. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 2016;35(16):1766–78.
Demarco B, Grayczyk JP, Bjanes E, Le Roy D, Tonnus W, Assenmacher CA, Radaelli E, Fettrelet T, Mack V, Linkermann A, Roger T, Brodsky IE, Chen KW, Broz P. Caspase-8-dependent gasdermin D cleavage promotes antimicrobial defense but confers susceptibility to TNF-induced lethality. Sci Adv. 2020 6(47):eabc3465. https://doi.org/10.1126/sciadv.abc3465
Chen KW, Demarco B, Heilig R, Shkarina K, Boettcher A, Farady CJ, Pelczar P, Broz P. Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. EMBO J. 2019;38(10):e101638.
Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, Brooks A, Xia S, Wu H, Kelliher MA, Berger SB, Gough PJ, Bertin J, Proulx MM, Goguen JD, Kayagaki N, Fitzgerald KA, Lien E. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science. 2018;362(6418):1064–9. https://doi.org/10.1126/science.aau2818.
Sarhan J, Liu BC, Muendlein HI, Li P, Nilson R, Tang AY, Rongvaux A, Bunnell SC, Shao F, Green DR, Poltorak A. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc Natl Acad Sci U S A. 2018;115(46):E10888–97. https://doi.org/10.1073/pnas.1809548115.
Sanjo H, Nakayama J, Yoshizawa T, Fehling HJ, Akira S, Taki S. Cutting edge: TAK1 safeguards macrophages against proinflammatory cell death. J Immunol. 2019;203(4):783–8. https://doi.org/10.4049/jimmunol.
Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, Shao F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547(7661):99–103. https://doi.org/10.1038/nature22393.
Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;3(8):14128. https://doi.org/10.1038/ncomms14128.
Hou J, Zhao R, Xia W, Chang CW, You Y, Hsu JM, Nie L, Chen Y, Wang YC, Liu C, Wang WJ, Wu Y, Ke B, Hsu JL, Huang K, Ye Z, Yang Y, Xia X, Li Y, Li CW, Shao B, Tainer JA, Hung MC. PD-L1-mediated gasdermin C expression switches apoptosis to pyroptosis in cancer cells and facilitates tumour necrosis. Nat Cell Biol. 2020;10:1264–75. https://doi.org/10.1038/s41556-020-0575-z.
Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (Gsdm) localizing to mouse chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mamm Genome. 2000;11(9):718–24. https://doi.org/10.1007/s003350010138.
Carl-McGrath S, Schneider-Stock R, Ebert M, Röcken C. Differential expression and localisation of gasdermin-like (GSDML), a novel member of the cancer-associated GSDMDC protein family, in neoplastic and non-neoplastic gastric, hepatic, and colon tissues. Pathology. 2008;40(1):13–24. https://doi.org/10.1080/00313020701716250.
Sun Q, Yang J, Xing G, Sun Q, Zhang L, He F. Expression of GSDML associates with tumor progression in uterine cervix cancer. Transl Oncol. 2008;1(2):73–83. https://doi.org/10.1593/tlo.08112.
Hergueta-Redondo M, Sarrió D, Molina-Crespo Á, Megias D, Mota A, Rojo-Sebastian A, García-Sanz P, Morales S, Abril S, Cano A, Peinado H, Moreno-Bueno G. Gasdermin-B promotes invasion and metastasis in breast cancer cells. PLoS One. 2014;9(3):e90099. https://doi.org/10.1371/journal.pone.0090099.
Watabe K, Ito A, Asada H, Endo Y, Kobayashi T, Nakamoto K, Itami S, Takao S, Shinomura Y, Aikou T, Yoshikawa K, Matsuzawa Y, Kitamura Y, Nojima H. Structure, expression and chromosome mapping of MLZE, a novel gene which is preferentially expressed in metastatic melanoma cells. Jpn J Cancer Res. 2001;92(2):140–51. https://doi.org/10.1111/j.1349-7006.2001.tb01076.x.
Saeki N, Usui T, Aoyagi K, Kim DH, Sato M, Mabuchi T, Yanagihara K, Ogawa K, Sakamoto H, Yoshida T, Sasaki H. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer. 2009;48(3):261–71. https://doi.org/10.1002/gcc.20636.
Miguchi M, Hinoi T, Shimomura M, Adachi T, Saito Y, Niitsu H, Kochi M, Sada H, Sotomaru Y, Ikenoue T, Shigeyasu K, Tanakaya K, Kitadai Y, Sentani K, Oue N, Yasui W, Ohdan H. Gasdermin C is upregulated by inactivation of transforming growth factor β receptor type II in the presence of mutated Apc, promoting colorectal cancer proliferation. PLoS One. 2016;11(11):e0166422. https://doi.org/10.1371/journal.pone.0166422.
Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, Junqueira C, Meza-Sosa KF, Mok TMY, Ansara J, Sengupta S, Yao Y, Wu H, Lieberman J. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579(7799):415–20. https://doi.org/10.1038/s41586-020-2071-9.
Zhou Z, He H, Wang K, Shi X, Wang Y, Su Y, Wang Y, Li D, Liu W, Zhang Y, Shen L, Han W, Shen L, Ding J, Shao F. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368(6494):7548. https://doi.org/10.1126/science.aaz7548.
Berkel C, Cacan E. Differential expression and copy number variation of gasdermin (GSDM) family members, pore-forming proteins in pyroptosis, in normal and malignant serous ovarian tissue. Inflammation. 2021. https://doi.org/10.1007/s10753-021-01493-0.
Kayagaki N, Kornfeld OS, Lee BL, Stowe IB, O’Rourke K, Li Q, Sandoval W, Yan D, Kang J, Xu M, Zhang J, Lee WP, McKenzie BS, Ulas G, Payandeh J, Roose-Girma M, Modrusan Z, Reja R, Sagolla M, Webster JD, Cho V, Andrews TD, Morris LX, Miosge LA, Goodnow CC, Bertram EM, Dixit VM. NINJ1 mediates plasma membrane rupture during lytic cell death. Nature. 2021;591(7848):131–6. https://doi.org/10.1038/s41586-021-03218-7.
Wang Y, Shao F. NINJ1, rupturing swollen membranes for cataclysmic cell lysis. Mol Cell. 2021;81(7):1370–1. https://doi.org/10.1016/j.molcel.2021.03.005.
Creekmore AL, Silkworth WT, Cimini D, Jensen RV, Roberts PC, Schmelz EM. Changes in gene expression and cellular architecture in an ovarian cancer progression model. PLoS One. 2011;6(3):e17676. https://doi.org/10.1371/journal.pone.0017676.
Li M, Balch C, Montgomery JS, Jeong M, Chung JH, Yan P, Huang TH, Kim S, Nephew KP. Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC Med Genomics. 2009;8(2):34. https://doi.org/10.1186/1755-8794-2-34.
Ganzfried BF, Riester M, Haibe-Kains B, Risch T, Tyekucheva S, Jazic I, Wang XV, Ahmadifar M, Birrer MJ, Parmigiani G, Huttenhower C, Waldron L. curatedOvarianData: clinically annotated data for the ovarian cancer transcriptome. Database (Oxford). 2013:bat013. https://doi.org/10.1093/database/bat013
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. https://doi.org/10.1038/nature10166.
Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–21. https://doi.org/10.1038/nmeth.3252.
Eckert MA, Coscia F, Chryplewicz A, Chang JW, Hernandez KM, Pan S, Tienda SM, Nahotko DA, Li G, Blaženović I, Lastra RR, Curtis M, Yamada SD, Perets R, McGregor SM, Andrade J, Fiehn O, Moellering RE, Mann M, Lengyel E (2019) Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 569(7758):723–728. https://doi.org/10.1038/s41586-019-1173-8
Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375(12):1109–12. https://doi.org/10.1056/NEJMp1607591.
Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, Scott AD, Krassowski M, Cherniack AD, Houlahan KE, Jayasinghe R, Wang LB, Zhou DC, Liu D, Cao S, Kim YW, Koire A, McMichael JF, Hucthagowder V, Kim TB, Hahn A, Wang C, McLellan MD, Al-Mulla F, Johnson KJ, Cancer Genome Atlas Research Network, Lichtarge O, Boutros PC, Raphael B, Lazar AJ, Zhang W, Wendl MC, Govindan R, Jain S, Wheeler D, Kulkarni S, Dipersio JF, Reimand J, Meric-Bernstam F, Chen K, Shmulevich I, Plon SE, Chen F, Ding L. Pathogenic germline variants in 10,389 Adult Cancers. Cell. 2018;173(2):355-370.e14. https://doi.org/10.1016/j.cell.2018.03.039.
Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V, Rao A, Schultz A, Li X, Sumazin P, Williams C, Mestdagh P, Gunaratne PH, Yau C, Bowlby R, Robertson AG, Tiezzi DG, Wang C, Cherniack AD, Godwin AK, Kuderer NM, Rader JS, Zuna RE, Sood AK, Lazar AJ, Ojesina AI, Adebamowo C, Adebamowo SN, Baggerly KA, Chen TW, Chiu HS, Lefever S, Liu L, MacKenzie K, Orsulic S, Roszik J, Shelley CS, Song Q, Vellano CP, Wentzensen N, Cancer Genome Atlas Research Network, Weinstein JN, Mills GB, Levine DA, Akbani R. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell. 2018;33(4):690-705.e9. https://doi.org/10.1016/j.ccell.2018.03.014.
Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11(1):6047. https://doi.org/10.1038/s41598-021-84787-5.
Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19(2):197–208. https://doi.org/10.1530/ERC-11-0329.
R Core Team (2020). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Wickham H, et al. Welcome to the tidyverse. J Open Source Softw. 2019;4(43):1686. https://doi.org/10.21105/joss.01686.
Hadley Wickham and Jennifer Bryan (2019). readxl: Read Excel Files. R package version 1.3.1. https://CRAN.R-project.org/package=readxl
Jeroen Ooms (2020). magick: advanced graphics and image-processing in R. R package version 2.5.2. https://CRAN.R-project.org/package=magick
Alboukadel Kassambara (2020). ggpubr: ‘ggplot2’ based publication ready plots. R package version 0.4.0. https://CRAN.R-project.org/package=ggpubr
Silge J, Robinson D (2016). tidytext: text mining and analysis using tidy DataPrinciples in R. _JOSS_, *1*(3). https://doi.org/10.21105/joss.00037https://doi.org/10.21105/joss.00037https://doi.org/10.21105/joss.00037>.
Claus O. Wilke (2020). ggtext: improved text rendering support for ‘ggplot2’. R package version 0.1.0. https://CRAN.R-project.org/package=ggtext
Jim Hester (2020). glue: interpreted string literals. R package version 1.4.2. https://CRAN.R-project.org/package=glue
JJ Allaire and Yihui Xie and Jonathan McPherson and Javier Luraschi and Kevin Ushey and Aron Atkins and Hadley Wickham and Joe Cheng and Winston Chang and Richard Iannone (2020). rmarkdown: dynamic documents for R. R package version 2.6. URL https://rmarkdown.rstudio.com.
Yihui Xie and Christophe Dervieux and Emily Riederer (2020). R Markdown Cookbook. Chapman and Hall/CRC. ISBN 9780367563837. URL https://bookdown.org/yihui/rmarkdown-cookbook.
Xie Y. knitr: a general-purpose package for dynamic report generation in R. R package version. 2020;1:30.
Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017. https://doi.org/10.1093/nar/gkx247.
Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36(3):1–48. https://doi.org/10.18637/jss.v036.i03.
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–14. https://doi.org/10.1093/nar/gkaa407.
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10. https://doi.org/10.1158/0008-5472.CAN-17-0307.
Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. https://doi.org/10.1186/s13059-016-1028-7.
Berkel C, Cacan E. Transcriptomic analysis reveals tumor stage- or grade-dependent expression of miRNAs in serous ovarian cancer. Hum Cell. 2021;34(3):862–77. https://doi.org/10.1007/s13577-021-00486-3.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Berkel, C., Cacan, E. Lower expression of NINJ1 (Ninjurin 1), a mediator of plasma membrane rupture, is associated with advanced disease and worse prognosis in serous ovarian cancer. Immunol Res 71, 15–28 (2023). https://doi.org/10.1007/s12026-022-09323-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-022-09323-7