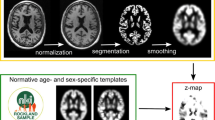
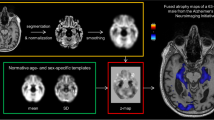
Abstract
The aim of this study was to assess whether the use of accelerated MRI scans in place of non-accelerated scans influenced brain volume and atrophy rate measures in controls and subjects with mild cognitive impairment and Alzheimer’s disease. We used data from 861 subjects at baseline, 573 subjects at 6 months and 384 subjects at 12 months from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). We calculated whole-brain, ventricular and hippocampal atrophy rates using the k-means boundary shift integral (BSI). Scan quality was visually assessed and the proportion of good quality accelerated and non-accelerated scans compared. We also compared MMSE scores, vascular burden and age between subjects with poor quality scans with those with good quality scans. Finally, we estimated sample size requirements for a hypothetical clinical trial when using atrophy rates from accelerated scans and non-accelerated scans. No significant differences in whole-brain, ventricular and hippocampal volumes and atrophy rates were found between accelerated and non-accelerated scans. Twice as many non-accelerated scan pairs suffered from at least some motion artefacts compared with accelerated scan pairs (p ≤ 0.001), which may influence the BSI. Subjects whose accelerated scans had significant motion had a higher mean vascular burden and age (p ≤ 0.05) whilst subjects whose non-accelerated scans had significant motion had poorer MMSE scores (p ≤ 0.05). No difference in estimated sample size requirements was found when using accelerated vs. non-accelerated scans. Accelerated scans reduce scan time and are better tolerated. Therefore it may be advantageous to use accelerated over non-accelerated scans in clinical trials that use ADNI-type protocols, especially in more cognitively impaired subjects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Volumetric magnetic resonance imaging (MRI) plays a key role in diagnosis, research and clinical trials in dementia. Single time-point structural MRI of the brain allows for the visualisation of atrophy patterns and quantification of brain volumes, which can aid diagnosis in neurodegenerative diseases. Volumes derived from a single MRI encapsulate prior atrophy but also a large amount of inter-subject variation. Serial MRI allows each individual to act as his or her own control meaning changes in volume can be visualised and measured. Such longitudinal measures have less variability and can be used to track progression allowing objective assessment of the effects of potentially disease-modifying therapies. This necessitates co-registration of the image series and for quantification, the application of techniques such as the boundary shift integral (BSI) to pairs of images (Fox and Freeborough 1997; Leung et al. 2010). Atrophy rates, as measured using the BSI have been shown to be sensitive biomarkers for Alzheimer’s disease (AD) and have been used as outcome measures in a number of clinical trials (Fox et al. 2005; Jack et al. 2008; Salloway et al. 2014). However, the quality of each scan in the series is important to provide robust and accurate results. Poor quality scans can be caused by patient-related factors such as movement during the scanning process.
A typical 3T 3D structural brain MRI with 1 mm resolution takes approximately 9 min to acquire but some subjects have difficulty remaining still for this time period, often resulting in motion artefacts rendering them unusable for quantification. Acquisition times can be reduced by employing parallel imaging techniques (e.g. with scan times of approximately 5 min), potentially reducing motion artefacts, scanning costs, as well as increasing patient comfort and compliance. Reducing scanning time may mean that a higher proportion of subjects recruited to a study or clinical trial has useable MRI scans.
However, accelerated acquisitions alter scan characteristics such as signal-to-noise ratio, noise distribution and tissue contrast. Before accelerated T1 scans can be used in place of non-accelerated T1 scans in clinical trials, it is essential that we understand how accelerated acquisitions may affect cross-sectional volumes and longitudinal atrophy rate measures. Previous studies have investigated the influence of using accelerated protocols in place of non-accelerated cross-sectionally on freesurfer morphometry measures (Wonderlick et al. 2009) and longitudinally on tensor based morphometry (TBM) based atrophy measures (Ching et al. 2015; Hua et al. 2016; Vemuri et al. 2015). Another study investigated the impact of changing from non-accelerated baseline scans to an accelerated follow-up scans on boundary shift integral (BSI) and deformation-based morphometry measures of atrophy (Leung et al. 2015). To date, however, there has been no study investigating the influence of using accelerated scans in place of non-accelerated at both baseline and follow-up, on the BSI, an important clinical trial outcome measure. TBM based atrophy measures and the BSI rely on different properties of MRI scans to quantify change necessitating assessment of the impact of using accelerated MRI scans on the BSI; the BSI tracks how much the surface of the brain (or structure of interest) has moved between baseline and follow-up by measuring changes in voxel intensities in a region around the brain boundary whereas TBM based measures use non-linear registration to align baseline and follow-up scans and volume change is estimated from the gradients of the resulting deformation fields. In addition, despite poor quality MRI scans routinely being excluded from studies (for reasons such as head coil failure, geometric distortions or patient motion), the question of whether subject characteristics differ between those who have useable MRI scans with those who have poor quality scans (due to patient motion) needs to be evaluated.
Therefore, the aims in this study were to: 1) compare whole brain, ventricular and hippocampal volumes at baseline and atrophy rates (measured using the BSI) over 6-month and 12-month intervals in accelerated and non-accelerated scans, 2) investigate whether there was a difference in the proportion of good quality accelerated and non-accelerated scans, 3) assess whether subject characteristics differed between subjects whose scans were considered unusable due to motion compared with those whose scans were of good quality and 4) compare estimated sample size requirements for a hypothetical clinical trial when using accelerated and non-accelerated scans.
Materials and Methods
Image Data and Acquisition
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). For up-to-date information, see www.adni-info.org. ADNI-GO and ADNI-2 included both accelerated and non-accelerated acquisition protocols for each participant. At the time of downloading baseline MRI scans were available for a total of 861 subjects, 6 month scans for 572 subjects, and 12 month scans for 384 subjects. All subjects used in the preparation of this article were scanned on 3T scanners and were new recruits to ADNI-GO/2. Image pre-processing included post-acquisition correction of gradient warping (Jovicich et al. 2006) and intensity non-uniformity correction using N3 (Sled et al. 1998). Scanners from three different manufacturers (Philips, Siemens and General Electric) were in use across the different sites. The three different scanner manufacturers use different acceleration protocols; details of the various MRI protocols are listed on the ADNI website (http://adni.loni.usc.edu/methods/documents/mri-protocols/). Non-accelerated scans were always acquired prior to the accelerated scans during the same scanning session. Subject demographics for all subjects with both accelerated and non-accelerated scans available at baseline, 6- and 12- months are shown in Table 1.
Quality Control (QC) of MRI Scans
First, imaging sites were instructed to immediately assess the quality of T1 weighted scans and to re-acquire if necessary; therefore in some cases more than one accelerated or non-accelerated T1 weighted scan was acquired in a particular session. QC was performed on all MRI scans at the Mayo Clinic before being released for download. This QC procedure entailed assessment of MRI scans for adherence to the ADNI scanning protocol, medical abnormalities and severe artefacts (such as metal-induced artefacts, head-coil failure etc.).
Only scans that passed this initial QC are available for download and so all analyses in this paper were only performed on the subset that passed initial QC by Mayo. At the Dementia Research Centre (UCL) further quality assessment of the MRI scans available to download was performed: individual scans were assessed visually for artefacts and very poor quality scans were excluded from further analysis (e.g. due to severe motion artefacts); once the scans were segmented and scan pairs registered (as detailed below), a single rater (EM), blinded to BSI values, assessed the quality of registered scan pairs. This assessment specifically looked at quality differences between the scans that may have affected the BSI measures. If a scan failed at this QC stage (from here on referred to as DRC QC), it was considered as failed for all BSI measures.
The BSI measures how much the boundary of a brain region has shifted between successive scans using normalised voxel intensities of co-registered images. Blurring due to patient motion can change the intensities of voxels at the brain boundary and may therefore render the BSI measure unreliable (Preboske et al. 2006). Since subjects had more opportunity to move during the longer non-accelerated scan we used a strict quality control process (i.e. erring on the side of excluding scans) as our first aim was to investigate whether the image acquisition, rather than differences in motion, might influence BSI measures. If motion, geometric distortions (due to different positioning in the scanner) or significant quality differences between baseline and follow-up scans were found, they were rated as unusable (for a more detailed description of the registration quality control process see Appendix). In addition, we excluded any subjects whose follow-up scans were performed on a different scanner from the baseline scans.
In order to test the reproducibility of the QC rating, the rater, blinded to diagnosis and scanner manufacturer, re-rated a sample of 60 scan pairs. The sample consisted scan pairs from 15 controls, 15 EMCI, 15 LMCI and 15 AD subjects. For each of the diagnostic groups 5 scan pairs were from GE scanners, 5 from Siemens scanners and 5 from Phillips scanners.
Quality Comparison of Accelerated and Non-accelerated Scans
To evaluate quality we examined the proportion of scan pairs that passed DRC QC. Patient motion is one of the major reasons a registered scan pair may fail but other reasons, such as geometric distortions due to positional differences may occur. We hypothesised that there may be more motion artefacts in non-accelerated scans than accelerated due to the longer time required for the non-accelerated scan, but that other reasons for failing QC such as geometric distortions due to positional differences would be independent of acquisition type. Therefore, in order to compare scan quality we excluded both the non-accelerated and accelerated scan pair if either were failed for reasons other than motion. Within the remaining scan pairs, we conducted a paired comparison of the proportion of accelerated scan pairs that passed DRC QC and the proportion of non-accelerated scan pairs that passed DRC QC using the McNemar test (McNemar 1947). Where more than one scan was acquired per session, we used the first scan to be acquired in this comparison (in the BSI comparisons we used the best scan acquired).
Segmentation and Volume Measurement
Brain regions were delineated by Multi-Atlas Propagation and Segmentation (MAPS) (Leung et al. 2011), visually checked and manually edited if necessary (edits included removing skull inclusions, spillages into non-brain tissue around the temporal lobes and cutting-off the brain stem at the most inferior slice of the cerebellum) . Segmentations were performed in native space. Ventricles (including the temporal horn and excluding the third and fourth ventricles) were segmented using a semi-automated technique. First the scans were registered to standard space (using a 9-dof-6 approach: 9 degrees of freedom (dof) registration from which the rigid body transform is extracted and applied to image). An upper threshold of 60% of the mean brain intensity was then applied to separate brain tissue from the cerebrospinal fluid. Manual editing was then performed to delineate the ventricular boundaries where required. For hippocampal segmentation, each baseline scan was first registered to standard space and the 6- and 12-month follow-up scans were then affinely registered to their baseline. Hippocampal regions were then automatically segmented using the Similarity and Truth Estimation for Propagated Segmentations (STEPS) algorithm (Cardoso et al. 2013) and were then manually checked and edited if necessary (e.g. editing in missing voxels around hippocampal cysts, removing the choroid plexus where it had been included, matching cut-offs in baseline and repeat scans). Left and right hippocampal volumes were summed. Differences in the numbers of scans with brain, ventricle and hippocampal volumes available were due to the workflow at the Dementia Research Centre, whereby scans are released for segmentation in batches and all regions are manually checked and edited where necessary. This means that not all available scans had hippocampal regions segmented and checked at the time of writing. We chose to include all available scans to maximise the numbers for the analyses.
Volume Change Measurement
For whole brain volume change, the 6- and 12-month scans were registered to the baseline scans using affine registration (12dof) and differential bias correction was applied. Likewise, for measuring ventricular volume change, the 6 and 12-month scans were registered to the baseline scans (which were registered to standard space prior to segmentation) using affine registration (12dof). Local 6-dof registration was performed separately for left and right side hippocampi after segmentation using the hippocampal regions dilated by 2 voxels. Volume change between follow-up and baseline was calculated using the robust boundary shift integral (KN-BSI) (Leung et al. 2010) for the whole-brain, using the fixed window BSI for the ventricles and using the double window BSI for the hippocampi (Leung et al. 2010). Left and right hippocampal atrophy rates were summed.
Comparison of Accelerated and Non-accelerated Volumes and Atrophy Rates
We used paired t-tests to compare baseline mean volumes between accelerated and non-accelerated scans and compared variance between accelerated and non-accelerated scans using Pitman’s test for equality of variance in paired samples. All statistical tests were performed in Stata 14.
To compare atrophy rates between accelerated and non-accelerated scan pairs we used a family of linear mixed models developed for the analysis of repeated measures of direct change (Frost et al. 2004). We only included scans that passed DRC QC in this analysis. These models account for the correlation between repeated atrophy measures and permit inclusion of all available atrophy measures in the analysis under the assumption that missing values are missing at random. The dependent variables in separate models were the ml loss of brains, ventricles and hippocampi as calculated by the BSI (see equation below). Time (years) between baseline and follow-up scans was included as a fixed-effect. Interactions terms between scan type and time were included to allow atrophy rate to vary with scan type. Random effects for visit were included to allow participants to have visit specific deviations from a linear trajectory, and random effect for time allow for participant specific differences from the mean rate of atrophy. The random effect of time was allowed to differ by diagnostic group to allow for differing between subject heterogeneity in atrophy rate. The model can be written as Eq. (1):
Where y ijk is the measured change between the j-th and k-th visit for the i-th individual, t ijk is the time interval between visits j and k, scantype is a categorical variable representing scan type (0 if accelerated, 1 if non-accelerated), EMCI is a categorical variable representing EMCI status (0 if not diagnosed as EMCI, 1 if diagnosed as EMCI) and likewise for LMCI and AD, β 0 is the mean atrophy rate in accelerated scans in controls, β 1 is the difference in mean atrophy rate between accelerated and non-accelerated scans and β 2 , β 3, β 4 are the fixed effects coefficients corresponding to a diagnosis of EMCI, LMCI or AD, b i terms are the random effect slope for subject i in the relevant diagnostic group and ε a,ijk and ε na,ijk are the error terms for accelerated and non-accelerated scans respectively.
We hypothesised the residual error (ε ijk ) may differ between non-accelerated scans and accelerated scans, as this represents the measurement error introduced in making the direct BSI measurement of atrophy. It was assumed that all other variance components, such as between subject heterogeneity in their atrophy rates could not be plausibly influenced by the scan type. Therefore, in order to test for differences in variance between accelerated and non-accelerated scans we used the likelihood ratio test on nested models, differing only in that one model specified with common residual variance (ε ijk ) and one model had separate residual variances by scan type (ε a , ijk ; ε na ,ijk ).
Comparison of Subject Characteristics between Passed and Failed Scans
We compared baseline MMSE, age and vascular burden- white matter hyperintensity (WMH) volumes (segmented on the FLAIR scans at UC Davis using an automated technique) (DeCarli et al. 1999) in subjects with scan pairs that failed QC (where one or both of the 0–6 and 0–12 month scan pairs failed DRC QC due to motion) with those who had only passed scan pairs (either 0–6 or 0–12 months or both if available) using linear regression separately by scan type (accelerated or non-accelerated). For the MMSE score comparison we adjusted for age, gender, years of education and diagnosis. For the WMH volume comparison we adjusted for age, baseline diagnosis and head size. Since WMH volumes and MMSE scores are not normally distributed we used bootstrapping (with 2000 iterations) to calculate bias-corrected and accelerated 95% confidence intervals (CI) for both of these analyses. For the age comparison we adjusted for baseline diagnosis, as the AD subjects were older than the EMCI subjects. We also compared the proportion of subjects with cognitive impairment (EMCI, LMCI or AD) between the groups (failed vs non-failed scans) using logistic regression and calculated odds ratios.
Sample Size Estimates
We estimated sample size requirements for a clinical trial to measure a 25% reduction in whole-brain atrophy rates in accelerated and non-accelerated scans using the standard formula: sample size per arm = (u + v)2 × (σ12 + σ22)/(μ1 − μ2)2 where u = 0.84 to provide 80% power and v = 1.96 to test at the 5% level; μ and σ are the mean and SDs of rates of atrophy in the treatment and placebo groups (assumes σ1 ≈ σ2). In order to directly compare the sample size requirements using accelerated and non-accelerated scans we included all 0–6 and 0–12 month scan pairs, whether or not they failed DRC QC.
Results
Quality Control
The majority of scans failed by Mayo were failed due to non-adherence to the ADNI protocol, incomplete coverage of the brain, metal induced artefacts or pathology. Only one subject had scans failed by Mayo due to severe motion. A total of 861 subjects had baseline accelerated and non-accelerated scans available to download from LONI. Of these 840 subjects had both a baseline accelerated and non-accelerated scan that passed DRC first pass QC. See Fig. 1 for a detailed breakdown of the number of subjects at each stage of the DRC QC.
Of the 573 subjects who had scans available at 6 months, scan pairs from 29 subjects were rated as unusable due to geometrical distortions due to positioning differencing in the scanner being detected on either or both of the accelerated and non-accelerated scans. A further 138 subjects had either an accelerated or non-accelerated scan pair rated as unusable due to motion. Of the 384 subjects who had scans available at 12 months, geometric distortion was detected on either the accelerated or non-accelerated scan pairs or both for 5 subjects and were therefore rated as unusable and a further 99 subjects had either an accelerated or non-accelerated scan pair rated as unusable due to motion. Examples of scan pairs that were failed due to motion are shown in Appendix 1.
In the sample of 60 scan pairs that were re-rated for reliability purposes there was 95% agreement between the initial and repeat ratings (Cohen’s kappa coefficient: 0.74).
Scan Quality Differences between Accelerated and Non-accelerated T1 MRI Scans
At both 6- and 12- month intervals, significantly more (twice as many) non-accelerated than accelerated scan pairs failed quality control due to motion artefact (in one or other of the scan pairs); at 6 months 14% of non-accelerated scan pairs were failed due to motion vs 7% of accelerated scan pairs (p < 0.001); at 12 months 20% of non-accelerated scan pairs failed due to motion vs 9% of accelerated scan pairs (p < 0.001).
Baseline Volumes
Table 2 summarises the mean baseline brain, ventricular and hippocampal volumes (summed left and right) measured from the baseline MRI scans. There were no differences in the mean baseline brain or hippocampal volumes measured using accelerated scans as compared with non-accelerated scans. The measured baseline ventricular volume was on average 0.09 ml lower for non-accelerated compared to accelerated scans. Although this difference was statistically significant (p < 0.001) it was very small at only 0.2% of the mean ventricular volume. There were no significant differences in variances of any of the measures.
Atrophy Rates
The results of the comparison of annualised brain, ventricular and hippocampal atrophy rates calculated using the BSI from accelerated and non-accelerated MRI scan pairs are shown in Table 3.
No significant differences in whole-brain, ventricular or hippocampal atrophy rates were found in any of the comparisons. Residual errors were generally slightly higher for non-accelerated scans, although the difference only reached statistical significance in the hippocampal comparison in Philips scanners.
Comparison of Subject Characteristics between those with Failed Scan Pairs Due to Motion and those with Passed Scan Pairs
Unadjusted baseline WMH volumes, age and MMSE scores are shown in Table 4. The results have been dichotomised by scan quality; those whose 0–6 & 0–12 months scan pairs both passed QC (if available) and those who had one or both of the 0–6 and 0–12 month scan pairs fail QC. The results of the regression analyses comparing these variables between these two subject groups are also shown.
In Accelerated Scan Pairs
Subjects who had a failed 0–6 or 0–12 month accelerated scan pair (due to motion) had a lower mean adjusted WMH volume (adjusted for age, diagnosis and head-size) and had a higher mean adjusted age (adjusted for diagnosis). There was no significant association between the proportion of cognitively impaired subjects and QC failure due to motion, nor was an association found between baseline MMSE score and QC scan pair failure.
In Non-accelerated Scan Pairs
A higher proportion (11% higher) of subjects who had a failed 0–6 or 0–12 month non-accelerated scan pair (due to motion) were cognitively impaired (with a diagnosis of MCI or AD) compared with those whose scan pairs passed QC (p = 0.01). Subjects who had a failed 0–6 or 0–12 month non-accelerated scan pair (due to motion) also had a lower mean adjusted MMSE score (adjusted for age, gender, education and diagnosis) (p ≤ 0.05). No significant difference in WMH volumes or age was found.
Sample Size Estimates
Mean annualised brain atrophy rates by diagnostic group are shown in Table 5 and estimated sample sizes for subjects with LMCI and AD are shown in Table 6. No significant differences in sample size requirements were found when comparing accelerated and non-accelerated scan pairs for any of the atrophy measures.
Discussion
We have shown that using an accelerated scan protocol, which reduces acquisition time compared with non-accelerated protocols, results in fewer scan pairs being excluded from subsequent analysis due to motion artefacts (which may affect the BSI) without significantly altering the absolute rates of change or sample sizes required for clinical trials.
The differences, adjusted for diagnosis, varied by scanner manufacturer. Differences in atrophy rates calculated from accelerated and non-accelerated scans from GE scanners were higher than in Siemens and Philips scanners. Residual errors were in general higher in non-accelerated scans, although the difference only reached statistical significance in the hippocampal comparison in Philips scanners. We also found differences in the characteristics of those who failed the accelerated and non-accelerated scan pairs: those who had failed non-accelerated scan pairs had lower MMSEs at baseline (more severe) and a higher proportion of them were cognitively impaired; whereas those whose accelerated scan pairs failed DRC QC were older and had greater WMH volumes. The difference in WMH volumes remained after adjustment for age. This may be the result of scan order: accelerated scans were always performed after non-accelerated scans and it could be that subjects with higher WMH volumes were unable to remain still for both scans (the non-accelerated scan as well as the accelerated scan) or it may suggest that subjects with more extensive white matter damage are less able to remain sufficiently motionless even at the shorter scanning times. Regardless, disregarding data from subjects with unusable scans due to motion changes the characteristics of the participants included in any analysis, meaning that they may have different characteristics from those who were originally recruited to the study.
One prior study investigated the influence of changing from a non-accelerated scanning protocol at the baseline to an accelerated protocol at repeat on the BSI and deformation-based morphometry atrophy measures (Leung et al. 2015). They found that the extent to which this change in protocol influenced atrophy measures depended on manufacturer, with little effect for some acquisitions and manufacturers. Another previous study investigated the influence of parallel imaging acquisition on brain volume measurements in 4 healthy control subjects (Krueger et al. 2012). They acquired 12 MPRAGE volumes in each session with a range of acceleration factors. They then repeated the same sequences 8 weeks later in each of the 4 subjects and found no significant differences in BSI measures even at high acceleration factors.
A further three previous studies investigated the influence of using accelerated T1 acquisitions on different measures of brain atrophy rate (Ching et al. 2015; Hua et al. 2016; Vemuri et al. 2015). These studies all used data from ADNI. One study (Ching et al. 2015) and it’s extension (Hua et al. 2016) found that measures of atrophy rate derived from tensor based morphometry were very similar between accelerated and non-accelerated acquisitions. Another study (Vemuri et al. 2015), which used a different atrophy rate measure based on symmetric diffeomorphic image normalisation and tensor based morphometry (TBM-Syn), did find some significant differences, with accelerated scan pairs tending to show higher TBM-Syn scores (or lower rates of atrophy) compared with non-accelerated pairs. Although previous studies have compared accelerated and non-accelerated scans for measures of atrophy based on TBM, different measures of atrophy are based on different properties of MRI scans and it may be that some measures are more affected by using accelerated protocols than others. For instance, it may be that TBM-Syn measures are more sensitive to differences in accelerated and non-accelerated scans (although the differences found in that study may have also been due to the more liberal inclusion of scans with motion than in this study). This is the first study to date to examine the impact of using accelerated scans in place of non-accelerated scans at both baseline and follow-up on the BSI – a measure used as a secondary outcome in clinical trials of treatments in AD. In addition, we investigated the influence of scanner manufacturer on atrophy rate outcomes as well as the demographics and vascular burden of those who had good quality vs. poor quality serial images, which adds to the existing literature.
There are some limitations to this study. First, subjects always had the non-accelerated scan prior to the accelerated scan. It could be that the finding of more motion artefacts in the non-accelerated scans was due to the ordering of the scans rather than the longer scan time. Secondly, the motion artefacts were visually rated and the rater was not blinded to the type of scan performed, which may have introduced some bias. We performed considered QC in order to be able to test the specific hypotheses of interest. However, it may be that had another rater performed this QC, different results may have been obtained. We did not assess rescans as part of the quality comparison as we wished to assess how quality was affected in the first scan of each type obtained. It may be that assessing rescans in place of original scans may have changed the results presented. Thirdly, given the importance of head coils on the quality of scans, it would be interesting to investigate the influence of head coil type on the differences between accelerated and non-accelerated scans. Unfortunately details of head coils used were not available for a large number of the ADNI subjects, so we were unable to perform this analysis in this study. Further, although our results indicate that atrophy rates measured using the BSI in accelerated scans are not markedly different from those measured in non-accelerated scans, it may be that different techniques for measuring brain atrophy rates are more susceptible to the changes in scan characteristics introduced by parallel imaging techniques. Notably, the differences in hippocampal atrophy rates between accelerated and non-accelerated scans were relatively higher than the differences in whole-brain and ventricular atrophy rates (~30% difference in hippocampal atrophy rates between accelerated and non-accelerated scans on Philips and GE scanners and ~15% difference on Siemens scanners) although the differences were not statistically significant. The SNR ratios in accelerated scans are highest near the head coils, on the surface of the brain and get progressively worse, as the distance from the coils increases (Krueger et al. 2012). The hippocampal BSI may therefore be more sensitive to the differences in accelerated and non-accelerated scans due to their location deep in the brain. It may be that with longer time intervals, these differences become more apparent and further studies would be required to investigate this.
In summary, the shorter scan time of accelerated scans may reduce the proportion of first-acquisition scans affected by motion artefacts. Therefore it may be advantageous to use accelerated T1 MRI scans rather than non-accelerated scans for assessing brain volumes and atrophy rates (BSI) in clinical trials. Importantly, the use of accelerated T1 structural MRI scans in place of non-accelerated scans does not appear to have an impact on whole-brain, ventricular and hippocampal volume and atrophy rate (BSI) measures. Finally, differences in subject characteristics were observed in those subjects whose scan pairs passed DRC QC from those whose scan pairs failed DRC QC due to motion in both accelerated and non-accelerated scans. Disregarding data from subjects who are unable to keep sufficiently still during an MRI scan to produce quality data ultimately biases the characteristics of the subject group in some way, possibly excluding those subjects who are more severe or have a more vascular form of the disease. However, the use of accelerated T1 volumetric scans rather than non-accelerated T1 volumetric scans may mean that higher quality longitudinal images can be obtained on a larger proportion of the original study population.
Information Sharing Statement
Data used in the preparation of this article were obtained from the Alzheimer’s disease Neuroimaging Initiative (ADNI, RRID:SCR_003007) database, which is available at adni.loni.usc.edu. In-house tools were used to generate the whole-brain, ventricular and hippocampal volumes and BSI measures and are available for download from adni.loni.usc.edu. Stata 14 was used for all statistical comparisons, available for purchase from www.stata.com/.
References
Cardoso, M. J., Leung, K., Modat, M., Keihaninejad, S., Cash, D., Barnes, J., et al. (2013). STEPS: similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcelation. Med Image Anal, 17(6), 671–684. doi:10.1016/j.media.2013.02.006.
Ching, C. R. K., Hua, X., Hibar, D. P., Ward, C. P., Gunter, J. L., Bernstein, M. A., et al. (2015). Does MRI scan acceleration affect power to track brain change? Neurobiol Aging, 36(Suppl 1), S167–S177. doi:10.1016/j.neurobiolaging.2014.05.039.
DeCarli, C., Miller, B. L., Swan, G. E., Reed, T., Wolf, P. A., Garner, J., et al. (1999). Predictors of brain morphology for the men of the NHLBI twin study. Stroke; a journal of cerebral circulation, 30(3), 529–536. doi:10.1161/01.STR.30.3.529.
Fox, N. C., & Freeborough, P. A. (1997). Brain atrophy progression measured from registered serial MRI: validation and application to Alzheimer’s disease. J Magn Reson Imaging, 7(6), 1069–1075. doi:10.1002/jmri.1880070620.
Fox, N. C., Black, R. S., Gilman, S., Rossor, M. N., Griffith, S. G., Jenkins, L., & Koller, M. (2005). Effects of Abeta immunization (AN1792) on MRI measures of cerebral volume in Alzheimer disease. Neurology, 64(9), 1563–1572. doi:10.1212/01.WNL.0000159743.08996.99.
Frost, C., Kenward, M. G., & Fox, N. C. (2004). The analysis of repeated “direct” measures of change illustrated with an application in longitudinal imaging. Stat Med, 23(21), 3275–3286. doi:10.1002/sim.1909.
Hua, X., Ching, C. R. K., Mezher, A., Gutman, B. A., Hibar, D. P., Bhatt, P., et al. (2016). MRI-based brain atrophy rates in ADNI phase 2: acceleration and enrichment considerations for clinical trials. Neurobiol Aging, 37, 26–37. doi:10.1016/j.neurobiolaging.2015.09.018.
Jack, C. R., Petersen, R. C., Grundman, M., Jin, S., Gamst, A., Ward, C. P., et al. (2008). Longitudinal MRI findings from the vitamin E and donepezil treatment study for MCI. Neurobiol Aging, 29(9), 1285–1295. doi:10.1016/j.neurobiolaging.2007.03.004.
Jovicich, J., Czanner, S., Greve, D., Haley, E., van der Kouwe, A., Gollub, R., et al. (2006). Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage, 30(2), 436–443. doi:10.1016/j.neuroimage.2005.09.046.
Krueger, G., Granziera, C., Jack, C. R., Gunter, J. L., Littmann, A., Mortamet, B., et al. (2012). Effects of MRI scan acceleration on brain volume measurement consistency. Journal of Magnetic Resonance Imaging: JMRI, 36(5), 1234–1240. doi:10.1002/jmri.23694.
Leung, K. K., Clarkson, M. J., Bartlett, J. W., Clegg, S., Jack, C. R., Weiner, M. W., et al. (2010). Robust atrophy rate measurement in Alzheimer’s disease using multi-site serial MRI: tissue-specific intensity normalization and parameter selection. NeuroImage, 50(2), 516–523. doi:10.1016/j.neuroimage.2009.12.059.
Leung, K. K., Barnes, J., Modat, M., Ridgway, G. R., Bartlett, J. W., Fox, N. C., & Ourselin, S. (2011). Brain MAPS: an automated, accurate and robust brain extraction technique using a template library. NeuroImage, 55(3), 1091–1108. doi:10.1016/j.neuroimage.2010.12.067.
Leung, K. K., Malone, I. M., Ourselin, S., Gunter, J. L., Bernstein, M. A., Thompson, P. M., et al. (2015). Effects of changing from non-accelerated to accelerated MRI for follow-up in brain atrophy measurement. NeuroImage, 107, 46–53. doi:10.1016/j.neuroimage.2014.11.049.
McNemar, Q. (1947). Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika, 12(2), 153–157. doi:10.1007/BF02295996.
Preboske, G. M., Gunter, J. L., Ward, C. P., & Jack, C. R. (2006). Common MRI acquisition non-idealities significantly impact the output of the boundary shift integral method of measuring brain atrophy on serial MRI. NeuroImage, 30(4), 1196–1202. doi:10.1016/j.neuroimage.2005.10.049.
Salloway, S., Sperling, R., Fox, N. C., Blennow, K., Klunk, W., Raskind, M., et al. (2014). Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer’s disease. N Engl J Med, 370(4), 322–333. doi:10.1056/NEJMoa1304839.
Sled, J. G., Zijdenbos, A. P., & Evans, A. C. (1998). A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging, 17, 87–97. doi:10.1109/42.668698.
Vemuri, P., Senjem, M. L., Gunter, J. L., Lundt, E. S., Tosakulwong, N., Weigand, S. D., et al. (2015). Accelerated vs. unaccelerated serial MRI based TBM-SyN measurements for clinical trials in Alzheimer’s disease. NeuroImage, 113, 61–69. doi:10.1016/j.neuroimage.2015.03.026.
Wonderlick, J. S., Ziegler, D. A., Hosseini-Varnamkhasti, P., Locascio, J. J., Bakkour, A., van der Kouwe, A., et al. (2009). Reliability of MRI-derived cortical and subcortical morphometric measures: effects of pulse sequence, voxel geometry, and parallel imaging. NeuroImage, 44(4), 1324–1333. doi:10.1016/j.neuroimage.2008.10.037.
Acknowledgements
EM and JB are funded by Alzheimers Research UK. MJC receives funding from EPSRC (EP/H046410/1). JMS acknowledges the support of the NIHR Queen Square Dementia BRU, the NIHR UCL/H Biomedical Research Centre, Wolfson Foundation, EPSRC (EP/J020990/1), MRC (CSUB19166), ARUK (ARUK-Network 2012-6-ICE; ARUK-PG2014-1946), Brain Research Trust ((UCC14191) and European Union’s Horizon 2020 research and innovation programme (Grant 666992). NCF has an NIHR Senior Investigator award and also receives support from the Wolfson Foundation and the NIHR Biomedical Research Unit in Dementia at UCL. NCF’s research group has received payment for consultancy or conducting studies from AVID, Eli Lilly, GSK, Janssen, Novartis, Roche and Sano. NCF receives no personal compensation for the activities mentioned above. The Dementia Research Centre is supported by Alzheimer’s Research UK, Brain Research Trust, and The Wolfson Foundation. This work was supported by the NIHR Queen Square Dementia Biomedical Research Unit. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a Group/Institutional Author.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Electronic supplementary material
ESM 1
(PPSX 2406 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Manning, E.N., Leung, K.K., Nicholas, J.M. et al. A Comparison of Accelerated and Non-accelerated MRI Scans for Brain Volume and Boundary Shift Integral Measures of Volume Change: Evidence from the ADNI Dataset. Neuroinform 15, 215–226 (2017). https://doi.org/10.1007/s12021-017-9326-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-017-9326-0