Abstract
Objective
To construct a risk prediction model for assisted diagnosis of Diabetic Nephropathy (DN) using machine learning algorithms, and to validate it internally and externally.
Methods
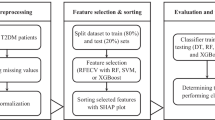
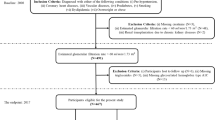
Firstly, the data was cleaned and enhanced, and was divided into training and test sets according to the 7:3 ratio. Then, the metrics related to DN were filtered by difference analysis, Least Absolute Shrinkage and Selection Operator (LASSO), Recursive Feature Elimination (RFE), and Max-relevance and Min-redundancy (MRMR) algorithms. Ten machine learning models were constructed based on the key variables. The best model was filtered by Receiver Operating Characteristic (ROC), Precision-Recall (PR), Accuracy, Matthews Correlation Coefficient (MCC), and Kappa, and was internally and externally validated. Based on the best model, an online platform had been constructed.
Results
15 key variables were selected, and among the 10 machine learning models, the Random Forest model achieved the best predictive performance. In the test set, the area under the ROC curve was 0.912, and in two external validation cohorts, the area under the ROC curve was 0.828 and 0.863, indicating excellent predictive and generalization abilities.
Conclusion
The model has a good predictive value and is expected to help in the early diagnosis and screening of clinical DN.





Similar content being viewed by others
Data availability
All data in this article can be found in the following databases: NPHDC, NHANES, and TWBB. An online platform has been created and you can access it through the following link (https://dn-prediction.shinyapps.io/DN-PRED-English).
References
M. Darenskaya, S. Kolesnikov, N. Semenova, L. Kolesnikova. Diabetic nephropathy: significance of determining oxidative stress and opportunities for antioxidant therapies. Int. J. Mol. Sci. 24 (2023). https://doi.org/10.3390/ijms241512378.
Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017, A systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1789–1858 (2018). https://doi.org/10.1016/s0140-6736(18)32279-7
M. Guedes, R. Pecoits-Filho, Can we cure diabetic kidney disease? Present and future perspectives from a nephrologist’s point of view. J. Intern. Med. 291, 165–180 (2022). https://doi.org/10.1111/joim.13424
Q. Hu, Y. Chen, X. Deng, Y. Li, X. Ma, J. Zeng, Y. Zhao, Diabetic nephropathy: Focusing on pathological signals, clinical treatment, and dietary regulation. Biomed. Pharmacother. 159, 114252 (2023). https://doi.org/10.1016/j.biopha.2023.114252
K. Zhang, Z. Fu, Y. Zhang, X. Chen, G. Cai, Q. Hong, The role of cellular crosstalk in the progression of diabetic nephropathy. Front. Endocrinol. (Lausanne) 14, 1173933 (2023). https://doi.org/10.3389/fendo.2023.1173933
M. Vučić Lovrenčić, S. Božičević, L. Smirčić Duvnjak, Diagnostic challenges of diabetic kidney disease. Biochem. Med. (Zagreb) 33, 030501 (2023). https://doi.org/10.11613/bm.2023.030501
R.Y. Choi, A.S. Coyner, J. Kalpathy-Cramer, M.F. Chiang, J.P. Campbell, Introduction to machine learning, neural networks, and deep learning. Transl. Vis. Sci. Technol. 9, 14 (2020). https://doi.org/10.1167/tvst.9.2.14
G.S. Handelman, H.K. Kok, R.V. Chandra, A.H. Razavi, M.J. Lee, H. Asadi, eDoctor: Machine learning and the future of medicine. J. Intern Med. 284, 603–619 (2018). https://doi.org/10.1111/joim.12822
R. Gupta, S. Kumari, A. Senapati, R.K. Ambasta, P. Kumar, New era of artificial intelligence and machine learning-based detection, diagnosis, and therapeutics in Parkinson’s disease. Ageing Res. Rev. 90, 102013 (2023). https://doi.org/10.1016/j.arr.2023.102013
Z. Bao, J. Bufton, R.J. Hickman, A. Aspuru-Guzik, P. Bannigan, C. Allen, Revolutionizing drug formulation development: The increasing impact of machine learning. Adv. Drug Deliv. Rev. 202, 115108 (2023). https://doi.org/10.1016/j.addr.2023.115108
J.B. Xue, S. Xia, X.Y. Wang, L.L. Huang, L.Y. Huang, Y.W. Hao, L.J. Zhang, S.Z. Li, Recognizing and monitoring infectious sources of schistosomiasis by developing deep learning models with high-resolution remote sensing images. Infect. Dis. Poverty 12, 6 (2023). https://doi.org/10.1186/s40249-023-01060-9
J.M. Yin, Y. Li, J.T. Xue, G.W. Zong, Z.Z. Fang, L. Zou, Explainable machine learning-based prediction model for diabetic nephropathy. J. Diabetes Res. 2024, 8857453 (2024). https://doi.org/10.1155/2024/8857453
M. Xu, H. Zhou, P. Hu, Y. Pan, S. Wang, L. Liu, X. Liu, Identification and validation of immune and oxidative stress-related diagnostic markers for diabetic nephropathy by WGCNA and machine learning. Front. Immunol. 14, 1084531 (2023). https://doi.org/10.3389/fimmu.2023.1084531
X.Z. Liu, M. Duan, H.D. Huang, Y. Zhang, T.Y. Xiang, W.C. Niu, B. Zhou, H.L. Wang, T.T. Zhang, Predicting diabetic kidney disease for type 2 diabetes mellitus by machine learning in the real world: A multicenter retrospective study. Front Endocrinol. (Lausanne) 14, 1184190 (2023). https://doi.org/10.3389/fendo.2023.1184190
S.M. Hosseini Sarkhosh, M. Hemmatabadi, A. Esteghamati, Development and validation of a risk score for diabetic kidney disease prediction in type 2 diabetes patients: a machine learning approach. J. Endocrinol. Invest 46, 415–423 (2023). https://doi.org/10.1007/s40618-022-01919-y
L. Zhao, H. Ren, J. Zhang, Y. Cao, Y. Wang, D. Meng, Y. Wu, R. Zhang, Y. Zou, H. Xu et al. Diabetic retinopathy, classified using the lesion-aware deep learning system, predicts diabetic end-stage renal disease in Chinese patients. Endocr. Pract. 26, 429–443 (2020). https://doi.org/10.4158/ep-2019-0512
C.T. Fan, J.C. Lin, C.H. Lee, Taiwan Biobank: a project aiming to aid Taiwan’s transition into a biomedical island. Pharmacogenomics 9, 235–246 (2008). https://doi.org/10.2217/14622416.9.2.235
S.v. Buuren. Flexible Imputation of Missing Data, 2nd edn. (Boca Raton, FL, 2018)
Z. Xu, D. Shen, Y. Kou, T. Nie. A synthetic minority oversampling technique based on Gaussian mixture model filtering for imbalanced data classification. IEEE Trans Neural Netw Learn Syst (2022). https://doi.org/10.1109/tnnls.2022.3197156
L. McInnes, J. Healy, J. Melville. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426 2018,
J. Chen, X. Zhang, D-MANOVA: fast distance-based multivariate analysis of variance for large-scale microbiome association studies. Bioinformatics 38, 286–288 (2021). https://doi.org/10.1093/bioinformatics/btab498
J.K. Tay, B. Narasimhan, T. Hastie. Elastic net regularization paths for all generalized linear models. J. Stat. Softw. 106 (2023). https://doi.org/10.18637/jss.v106.i01
Y. Han, L. Huang, F. Zhou, A dynamic recursive feature elimination framework (dRFE) to further refine a set of OMIC biomarkers. Bioinformatics 37, 2183–2189 (2021). https://doi.org/10.1093/bioinformatics/btab055
H. Peng, F. Long, C. Ding, Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 27, 1226–1238 (2005). https://doi.org/10.1109/tpami.2005.159
L. Breiman, Random forests. Mach. Learn. 45, 5–32 (2001). https://doi.org/10.1023/A:1010933404324
T. Chen, C. Guestrin. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining pp. 785–794 (2016)
E. Alfaro, M. Gáamez, N. García. adabag: An R package for classification with boosting and bagging. J. Stat. Softw. 2013, 54, https://doi.org/10.18637/jss.v054.i02
L. Prokhorenkova, G. Gusev, A. Vorobev, A.V. Dorogush, A. Gulin. CatBoost: unbiased boosting with categorical features. Adv. Neural Inform. Process. Syst. 31 (2018). https://doi.org/10.48550/arXiv.1706.09516
G. Ke, Q. Meng, T. Finley, T. Wang, W. Chen, W. Ma, Q. Ye, T.-Y. Liu. LightGBM: a highly efficient gradient boosting decision tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, California, USA, 2017; pp. 3149–3157
M. Abadi, P. Barham, J. Chen, Z. Chen, X. Zhang. TensorFlow: A system for large-scale machine learning. USENIX Association 2016, 265–283, https://doi.org/10.48550/arXiv.1605.08695
T.A. Dejenie, E.C. Abebe, M.A. Mengstie, M.A. Seid, N.A. Gebeyehu, G.A. Adella, G.A. Kassie, A.Y. Gebrekidan, M.M. Gesese, K.D. Tegegne et al. Dyslipidemia and serum cystatin C levels as biomarker of diabetic nephropathy in patients with type 2 diabetes mellitus. Front Endocrinol. (Lausanne) 14, 1124367 (2023). https://doi.org/10.3389/fendo.2023.1124367
A.K. Clift, D. Dodwell, S. Lord, S. Petrou, M. Brady, G.S. Collins, J. Hippisley-Cox, Development and internal-external validation of statistical and machine learning models for breast cancer prognostication: cohort study. Bmj 381, e073800 (2023). https://doi.org/10.1136/bmj-2022-073800
V. Subbiah, The next generation of evidence-based medicine. Nat. Med 29, 49–58 (2023). https://doi.org/10.1038/s41591-022-02160-z
R.D. Joshi, C.K. Dhakal. Predicting type 2 diabetes using logistic regression and machine learning approaches. Int. J. Environ. Res. Public Health 18 (2021). https://doi.org/10.3390/ijerph18147346
A. Zanchi, A.W. Jehle, F. Lamine, B. Vogt, C. Czerlau, S. Bilz, H. Seeger, S. de Seigneux, Diabetic kidney disease in type 2 diabetes: a consensus statement from the Swiss Societies of Diabetes and Nephrology. Swiss Med Wkly 153, 40004 (2023). https://doi.org/10.57187/smw.2023.40004
B.F. Palmer, Change in albuminuria as a surrogate endpoint for cardiovascular and renal outcomes in patients with diabetes. Diabetes Obes. Metab. 25, 1434–1443 (2023). https://doi.org/10.1111/dom.15030
X. Ren, N. Kang, X. Yu, X. Li, Y. Tang, J. Wu, Prevalence and association of diabetic nephropathy in newly diagnosed Chinese patients with diabetes in the Hebei province: A single-center case-control study. Medicine (Baltimore) 102, e32911 (2023). https://doi.org/10.1097/md.0000000000032911
S. Chen, L. Chen, H. Jiang, Prognosis and risk factors of chronic kidney disease progression in patients with diabetic kidney disease and non-diabetic kidney disease: a prospective cohort CKD-ROUTE study. Ren. Fail 44, 1309–1318 (2022). https://doi.org/10.1080/0886022x.2022.2106872
K. Azushima, J.P. Kovalik, T. Yamaji, J. Ching, T.W. Chng, J. Guo, J.J. Liu, M. Nguyen, R.B. Sakban, S.E. George, et al. Abnormal lactate metabolism is linked to albuminuria and kidney injury in diabetic nephropathy. Kidney Int. (2023). https://doi.org/10.1016/j.kint.2023.08.006
J.G. Greener, S.M. Kandathil, L. Moffat, D.T. Jones, A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 23, 40–55 (2022). https://doi.org/10.1038/s41580-021-00407-0
M. Garofolo, V. Napoli, D. Lucchesi, S. Accogli, M.L. Mazzeo, P. Rossi, E. Neri, S. Del Prato, G. Penno, Type 2 diabetes albuminuric and non-albuminuric phenotypes have different morphological and functional ultrasound features of diabetic kidney disease. Diabetes Metab. Res Rev. 39, e3585 (2023). https://doi.org/10.1002/dmrr.3585
Acknowledgements
We thank the National Population Health Data Center of China, the National Health and Nutrition Examination Survey of the United States, and Taiwan Biobank for providing data support.
Funding
This study was supported by the College Students’ Innovative Entrepreneurial Training Plan Program (202310367071).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Data collection and analysis were performed by J.J.M. The first draft of the manuscript was written by J.J.M., S.G.A., and M.H.C. The revision of the manuscript was completed by L.Z. and J.L. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, J., An, S., Cao, M. et al. Integrated machine learning and deep learning for predicting diabetic nephropathy model construction, validation, and interpretability. Endocrine (2024). https://doi.org/10.1007/s12020-024-03735-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12020-024-03735-1