Abstract
Background
Graves’ disease (GD) is an autoimmune disease, the incidence of which is increasing yearly. GD requires long-life therapy. Therefore, the potential immune-related biomarkers of GD need to be studied.
Method
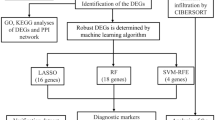
In our study, differentially expressed genes (DEGs) were derived from the online Gene Expression Omnibus (GEO) microarray expression dataset GSE71956. Protein‒protein interaction (PPI) network analyses were used to identify hub genes, which were validated by qPCR. GSEA was used to screen potential pathways and related immune cells. Next, CIBERSORT analysis was used to further explore the immune subtype distribution pattern among hub genes. ROC curves were used to analyze the specificity and sensitivity of hub genes.
Result
44 DEGs were screened from the GEO dataset. Two hub genes, EEF1A1 and EIF4B, were obtained from the PPI network and validated by qPCR (p < 0.05). GSEA was conducted to identify potential pathways and immune cells related to these the two hub genes. Immune cell subtype analysis revealed that hub genes had extensive associations with many different types of immune cells, particularly resting memory CD4+ T cells. AUCs of ROC analysis were 0.687 and 0.733 for EEF1A1 and EIF4B, respectively.
Conclusion
Our study revealed two hub genes, EEF1A1 and EIF4B, that are associated with resting memory CD4+ T cells and potential immune-related molecular biomarkers and therapeutic targets of GD.






Similar content being viewed by others
References
Y. He, et al. Association of HLA-B and HLA-DRB1 polymorphisms with antithyroid drug-induced agranulocytosis in a Han population from northern China. Sci. Rep. 7(1), 11950 (2017).
S. De Leo, S.Y. Lee, L.E. Braverman,, Hyperthyroidism. Lancet 388(10047), 906–918 (2016)
T.J. Smith, L. Hegedus, Graves’ disease. N. Engl. J. Med. 375(16), 1552–1565 (2016)
P.N. Taylor et al. Global epidemiology of hyperthyroidism and hypothyroidism. Nat. Rev. Endocrinol. 14(5), 301–316 (2018)
T.F. Davies et al. Graves’ disease. Nat. Rev. Dis. Prim. 6(1), 52 (2020)
H.T. Hao et al. Treatment of Graves’ ophthalmopathy with an in-house Phosphorus-32 source: Initial clinical observations. Exp. Ther. Med 14(4), 2795–2800 (2017)
W. Gu, S. Miller, C.Y. Chiu, Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev. Pathol. 14, 319–338 (2019)
A. Antonelli et al. Graves’ disease: Clinical manifestations, immune pathogenesis (cytokines and chemokines) and therapy. Best. Pr. Res Clin. Endocrinol. Metab. 34(1), 101388 (2020)
M. Limbach et al. Epigenetic profiling in CD4+ and CD8+ T cells from Graves’ disease patients reveals changes in genes associated with T cell receptor signaling. J. Autoimmun. 67, 46–56 (2016)
T.J. Smith, L. Hegedus, Graves’ disease. N. Engl. J. Med 376(2), 185 (2017)
J.Q. Chen, P. Szodoray, M. Zeher, Toll-like receptor pathways in autoimmune diseases. Clin. Rev. Allergy Immunol. 50(1), 1–17 (2016)
K.A. Dvornikova et al. Polymorphism of toll-like receptor genes and autoimmune endocrine diseases. Autoimmun. Rev. 19(4), 102496 (2020)
S.Y. Han et al. High-mobility group box 1 is associated with the inflammatory pathogenesis of graves’ orbitopathy. Thyroid 29(6), 868–878 (2019)
S. Shah et al. Ras and Rap1: A tale of two GTPases. Semin Cancer Biol. 54, 29–39 (2019)
N. Minato, K. Kometani, M. Hattori, Regulation of immune responses and hematopoiesis by the Rap1 signal. Adv. Immunol. 93, 229–264 (2007)
J.J. Hamey, M.R. Wilkins, Methylation of elongation factor 1A: where, who, and why?. Trends Biochem Sci. 43(3), 211–223 (2018)
J. Pelletier, N. Sonenberg, The organizing principles of eukaryotic ribosome recruitment. Annu Rev. Biochem 88, 307–335 (2019)
H.J. Lee et al. Immunogenetics of autoimmune thyroid diseases: A comprehensive review. J. Autoimmun. 64, 82–90 (2015)
P.S. Minhas et al. Macrophage de novo NAD(+) synthesis specifies immune function in aging and inflammation. Nat. Immunol. 20(1), 50–63 (2019)
E.L. Mills et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167(2), 457–470 e13 (2016)
S.A. Vardhana et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 21(9), 1022–1033 (2020)
Y. Zhao et al. m(6)A-binding proteins: the emerging crucial performers in epigenetics. J. Hematol. Oncol. 13(1), 35 (2020)
S. Fang et al. Mechanisms that underly T cell immunity in graves’ orbitopathy. Front Endocrinol. (Lausanne) 12, 648732 (2021)
S. Crotty, Do memory CD4 T cells keep their cell-type programming: plasticity versus fate commitment? Complexities of interpretation due to the heterogeneity of memory CD4 T cells, including T follicular helper cells. Cold Spring Harb. Perspect. Biol. 10(3), a032102 (2018)
S. Sengupta, R.F. Siliciano, Targeting the latent reservoir for HIV-1. Immunity 48(5), 872–895 (2018)
S. Sureshchandra et al. Phenotypic and epigenetic adaptations of cord blood CD4+ T cells to maternal obesity. Front Immunol. 12, 617592 (2021)
Z. Wang et al. IP-10 promotes latent HIV infection in resting memory CD4(+) T cells via LIMK-cofilin pathway. Front Immunol. 12, 656663 (2021)
N. Corcos et al. Oral Fc-coupled preproinsulin achieves systemic and thymic delivery through the neonatal Fc receptor and partially delays autoimmune diabetes. Front Immunol. 12, 616215 (2021)
T. Liu et al. Human eukaryotic elongation factor 1A forms oligomers through specific cysteine residues. Acta Biochim Biophys. Sin. (Shanghai) 47(12), 1011–1017 (2015)
Z. Cai et al. A three-gene signature and clinical outcome in pediatric acute myeloid leukemia. Clin. Transl. Oncol. 23(4), 866–873 (2021)
H.J. Ditzel et al. Cloning and expression of a novel human antibody-antigen pair associated with Felty’s syndrome. Proc. Natl Acad. Sci. USA 97(16), 9234–9239 (2000)
Q. Feng et al. Etodolac improves collagen induced rheumatoid arthritis in rats by inhibiting synovial inflammation, fibrosis and hyperplasia. Mol. Biomed. 2(1), 33 (2021)
Acknowledgements
Throughout the writing of this dissertation, I have received a great deal of support and assistance. I would first like to thank my supervisor, H.X., whose expertise was invaluable in formulating the research questions and methodology. Your insightful feedback pushed me to sharpen my thinking and brought my work to a higher level. I would particularly like to acknowledge my colleagues J.W., H.Z., B.L., Y.C., F.Q., and J.L. for their wonderful collaboration and patient support.
Funding
This work has been in part supported by the National Natural Science Foundation of China (81773741 and 81973329) and Natural Science Foundation Project of Shanghai (19ZR1440800).
Author information
Authors and Affiliations
Contributions
Y.Z., X.X. and H.X. conceived of and designed the project. J.W., B.L. and F.Q. acquired the data, and Y.C., B.L. and H.Z. analyzed and interpreted the data. Y.Z. and J.W. wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethical Committee of Shanghai General Hospital (Date 2018-2-27/No 2018KY088).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Wei, J., Zhou, H. et al. Identification of two potential immune-related biomarkers of Graves’ disease based on integrated bioinformatics analyses. Endocrine 78, 306–314 (2022). https://doi.org/10.1007/s12020-022-03156-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12020-022-03156-y