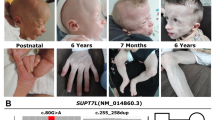
Abstract
A growing number of histone modifiers are involved in human neurodevelopmental disorders, suggesting that proper regulation of chromatin state is essential for the development of the central nervous system. Among them, heterozygous de novo variants in KMT2A, a gene coding for histone methyltransferase, have been associated with Wiedemann–Steiner syndrome (WSS), a rare developmental disorder mainly characterized by intellectual disability (ID) and hypertrichosis. As KMT2A is known to regulate the expression of multiple target genes through methylation of lysine 4 of histone 3 (H3K4me), we sought to investigate the transcriptomic consequences of KMT2A variants involved in WSS. Using fibroblasts from four WSS patients harboring loss-of-function KMT2A variants, we performed RNA sequencing and identified a number of genes for which transcription was altered in KMT2A-mutated cells compared to the control ones. Strikingly, analysis of the pathways and biological functions significantly deregulated between patients with WSS and healthy individuals revealed a number of processes predicted to be altered that are relevant for hypertrichosis and intellectual disability, the cardinal signs of this disease.



Similar content being viewed by others
References
Aggarwal, A., Rodriguez, D. F., & Northrup, H. (2017). Wiedemann-Steiner syndrome: Novel pathogenic variant and review of literature. European Journal of Medical Genetics, 60, 285–288.
Blake, J. A., et al. (2013). Gene Ontology annotations and resources. Nucleic Acids Research, 41(Database issue), D530–D535.
Brookes, E., et al. (2015). Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Human Molecular Genetics, 24(10), 2861–2872.
Chen, J., et al. (2005). Endothelial nitric oxide synthase regulates brain-derived neurotrophic factor expression and neurogenesis after stroke in mice. Journal of Neurosciences, 25(9), 2366–2375.
Chen, T., & Dent, S. Y. (2014). Chromatin modifiers and remodellers: regulators of cellular differentiation. Nature Review Genetics, 15(2), 93–106.
Cosgrove, M. S., & Patel, A. (2010). Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. FEBS Journal, 277(8), 1832–1842.
de Hoon, M. J., Imoto, S., Nolan, J., & Miyano, S. (2004). Open source clustering software. Bioinformatics, 20(9), 1453–1454.
Del Rizzo, P. A., & Trievel, R. C. (2011). Substrate and product specificities of SET domain methyltransferases. Epigenetics, 6(9), 1059–1067.
Diehl, F., et al. (2007). The histone methyltransferase MLL is an upstream regulator of endothelial-cell sprout formation. Blood, 109(4), 1472–1478.
Dobin, A., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29(1), 15–21.
Gao, J., et al. (2016). Transcription factor Six2 mediates the protection of GDNF on 6-OHDA lesioned dopaminergic neurons by regulating Smurf1 expression. Cell Death Disease, 7, e2217.
Guenther, M. G., et al. (2005). Global and Hox-specific roles for the MLL1 methyltransferase. Proceedings of the National Academy of Sciences USA, 102(24), 8603–8608.
Hellsten, Y., et al. (2012). Vasodilator interactions in skeletal muscle blood flow regulation. Journal of Physiology, 590(24), 6297–6305.
Herman, A., & Herman, A. P. (2016). Mechanism of action of herbs and their active constituents used in hair loss treatment. Fitoterapia, 114, 18–25.
Jones, W. D., et al. (2012). De novo mutations in MLL cause Wiedemann-Steiner syndrome. American Journal of Human Genetics, 91(2), 358–364.
Katada, S., & Sassone-Corsi, P. (2010). The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nature Structural & Molecular Biology, 17(12), 1414–1421.
Kerimoglu, C., et al. (2017). KMT2A and KMT2B mediate memory function by affecting distinct genomic regions. Cell Reports, 20(3), 538–548.
Kouzarides, T. (2007). Chromatin modifications and their function. Cell, 128(4), 693–705.
Lebrun, N., et al. (2017). Molecular and cellular issues of KMT2A variants involved in Wiedemann-Steiner syndrome. European Journal of Human Genetics, 26(1), 107–116.
Li, Z., et al. (2008). Inhibitory effect of D1-like and D3 dopamine receptors on norepinephrine-induced proliferation in vascular smooth muscle cells. American Journal of Physiology Heart and Circulation Physiology, 294(6), H2761–H2768.
Liao, Y., Smyth, G. K., & Shi, W. (2014). FeatureCounts: an efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930.
Lin, W. H., et al. (2015). Fibroblast growth factors stimulate hair growth through β-catenin and Shh expression in C57BL/6 mice. BioMed Research International, 2015, 730139.
Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15, 550.
Malik, S., & Bhaumik, S. R. (2010). Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS Journal, 277(8), 1805–1821.
Merzdorf, C. S., & Sive, H. L. (2006). The zic1 gene is an activator of Wnt signaling. The International Journal of Developmental Biology, 50(7), 611–617.
Milne, T. A., et al. (2005). Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proceedings of the National Academy of Sciences USA, 102(3),749–754.
Miyake, N., et al. (2016). Delineation of clinical features in Wiedemann-Steiner syndrome caused by KMT2A mutations. Clinical Genetics, 89(1), 115–119.
Mize, R. R., et al. (1998). The role of nitric oxide in development of the patch-cluster system and retinocollicular pathways in the rodent superior colliculus. Progress in Brain Research, 118, 133–152.
Ohuchi, H., et al. (2003). Fibroblast growth factor 10 is required for proper development of the mouse whiskers. Biochemical and Biophysical Research Communications, 302(3), 562–567.
Oyama, T., et al. (2013). Cleavage of TFIIA by Taspase1 activates TRF2-specified mammalian male germ cell programs. Developmental Cell, 27(2), 188–200.
Parkel, S., Lopez-Atalaya, J. P., & Barco, A. (2013). Histone H3 lysine methylation in cognition and intellectual disability disorders. Learning & Memory, 20(10), 570–579.
Pezzani, L., Milani, D., & Tadini, G. (2015). Intellectual disability: When the hypertrichosis is a clue. Journal of Pediatrics Genetics, 4(3), 154–158.
Pontén, F., Jirström, K., & Uhlen, M. (2008). The Human protein atlas—a tool for pathology. Journal of Pathology, 216(4), 387–393.
Pourebrahim, R., et al. (2007). ZIC1 gene expression is controlled by DNA and histone methylation in mesenchymal proliferations. FEBS Letters, 581(26), 5122–5126.
Powell, B. C., et al. (1998). The Notch signalling pathway in hair growth. Mechanisms of Development, 78(1–2), 189–192.
Rishikaysh, P., et al. (2014). Signaling involved in hair follicle morphogenesis and development. International Journal of Molecular Sciences, 15(1), 1647–1670.
Ronan, J. L., Wu, W., & Crabtree, G. R. (2013). From neural development to cognition: unexpected roles for chromatin. Nature Reviews Genetics, 14(5), 347–359.
Rosenbloom, K. R., et al. (2010). ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Research, 38(Database issue), D620–D625.
Saldanha, A. J. (2004). Java TreeView–extensible visualization of microarray data. Bioinformatics, 20(17), 3246–3248.
Subramanian, A., et al (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America, 102(43), 15545–15550.
Sun, Y., et al. (2017). Further delineation of the phenotype of truncating KMT2A mutations: The extended Wiedemann-Steiner syndrome. American Journal of Medical Genetics A, 173(2), 510–514.
Twigg, S. R., et al. (2015). Gain-of-Function Mutations in ZIC1 Are Associated with Coronal Craniosynostosis and Learning Disability. American Journal of Human Genetics, 97(3), 378–388.
Vallianatos, C. N., & Iwase, S. (2015). Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics, 7(3), 503–519.
Wang, D., et al. (2013). MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nature Cell Biology, 15(10), 1153–1163.
Wang, X., & Zhang, X. (2011). Pinpointing transcription factor binding sites from ChIP-seq data with SeqSite. BMC Systems Biology, 5, S3.
Wiersma, M., et al. (2016). Protein kinase Msk1 physically and functionally interacts with the KMT2A/MLL1methyltransferase complex and contributes to the regulation of multiple target genes. Epigenetics & Chromatin, 9, 52.
Zhang, P., Bergamin, E., & Couture, J. F. (2013). The many facets of MLL1 regulation. Biopolymers, 99(2), 136–145.
Acknowledgements
We thank the families for their enthusiastic participation. We thank Patrick Nusbaum and Arnaud Hubas for providing primary cultures of fibroblasts. We also thank Franck Letourneur (Genomics Platform at the Institut Cochin, Paris, France) for assistance with RNA sequencing and analysis. This work was supported by the Université Paris Descartes and the Labex “Who I am?”
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflicting interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
12017_2018_8502_MOESM1_ESM.docx
Figure S1: Genomic positions of primers used for chromatin immunoprecipitation assays: ChIP-Seq data of NHDF-Ad (adult dermal fibroblasts) H3K4me3 Histone (from https://www.encodeproject.org/experiments/ENCSR000APR/) (Rosenbloom et al. 2010) were used to design ChIP-qPCR primers. Mapping of amplicons (”Your Seq”) was represented for the different H3K4Me3 target regions. (DOCX 87 KB)
12017_2018_8502_MOESM2_ESM.pptx
Figure S2: PCA plot for control (Te03, Te04, Te05, F298 and F157) (green) and WSS samples (F70274, F66867, F66262 and LEB) (red). (PPTX 409 KB)
12017_2018_8502_MOESM3_ESM.pptx
Figure S3: An MA-Plot of changes due to KMT2A pathogenic variant. The log2 fold change is plotted on the y-axis and the average of the normalized counts is shown on the x-axis. Each gene is represented with a dot. DEGs are shown in red. (PPTX 983 KB)
12017_2018_8502_MOESM4_ESM.pptx
Figure S4. Validation of RNA-Seq data by qRT-PCR. Correlation plot of log2 fold changes obtained by RNA-Seq (x-axis) and by RT-qPCR (y-axis) for selected genes with transcriptional changes (black dots). Direction of change correlates fully with r2 for a linear regression model at 0.85. Black line represents the linear regression line and r2 represents the correlation coefficient. Correlation obtained by RNA-Seq and qRT-PCR using the primary samples (on top) and correlation obtained by RNA-Seq and qRT-PCR using other independent fibroblast cultures (on bottom). (PPTX 88 KB)
12017_2018_8502_MOESM6_ESM.xls
Table S2: Differentially expressed genes (DEG)s with a p value <0.05 identified from RNA-Seq. by RNA-Seq in fibroblasts from WSS patients sorted by p value. (XLS 198 KB)
12017_2018_8502_MOESM7_ESM.xlsx
Table S3: Differentially expressed genes (DEG)s with a p value <0.01 and a Fold Change >2 (with > 10 reads) identified by RNA-Seq in fibroblasts from WSS patients sorted by p value. (XLSX 24 KB)
12017_2018_8502_MOESM8_ESM.xls
Table S4: Top 10 of the differentially expressed genes (DEG)s (down-regulated genes in grey and up-regulated genes in white) exhibiting the higher fold changes. (XLS 32 KB)
12017_2018_8502_MOESM9_ESM.xlsx
Table S5: Top 20 of the differentially expressed genes (DEG)s (down-regulated genes in grey and up-regulated genes in white) exhibiting the strongest p values. (XLSX 11 KB)
12017_2018_8502_MOESM10_ESM.xlsx
Table S6: mRNA expression estimated by number of KMT2A normalized reads obtained by RNA sequencing in controls and in KMT2A mutated patients (in yellow). (XLSX 8 KB)
12017_2018_8502_MOESM11_ESM.xlsx
Table S7: List of differentially expressed genes assessed by quantitative reverse transcription – PCR. Log2 Fold change results obtained in RNA-Seq samples and in replicated samples are indicated for each analyzed gene. (XLSX 10 KB)
12017_2018_8502_MOESM12_ESM.xls
Table S8: Canonical pathways enriched in DEGs sorted by p value of enrichment (nd = non-determined)(-log(p value)=1.3 ~ p value=0.05)). Using the IPA software in the GO database (http://www.geneontology.org/) (Blake et al. 2013), the physiological functions and biological processes of the DEGs were classified into the different categories. Notably, some DEGs were significantly enriched in the eNOS signaling, and Axonal Guidance Signaling pathway. (XLS 25 KB)
12017_2018_8502_MOESM13_ESM.xlsx
Table S9: Upstream analysis (Activated/inhibited diseases and functions enriched in DEGs) using the list of DEGs with a p value <0.05 and a minimum fold change of 1.2 (and with more than 10 reads) showing that KMT2A activity was predicted to be inhibited. (XLSX 8 KB)
12017_2018_8502_MOESM14_ESM.xls
Table S10: Significantly deregulated IPA-networks using the list of DEGs with a p value <0.01 and a fold change >2. Network analysis showed string association of modulated gene expressions with skin and brain networks, represented by high network scores. (XLS 29 KB)
12017_2018_8502_MOESM15_ESM.xlsx
Table S11: Top 10 of diseases and functions (with a positive activation z score) enriched in genes using the list of DEGs with a p value <0.01, showing an overrepresentation of hyperplasia of skin (positive activation z score) (XLSX 10 KB)
12017_2018_8502_MOESM16_ESM.xlsx
Table S12: Top 10 of diseases and functions (with a negative activation z score) enriched in genes using the list of DEGs with a p value <0.01, showing an overrepresentation of brain functions (negative activation z score). (XLSX 10 KB)
Rights and permissions
About this article
Cite this article
Mietton, L., Lebrun, N., Giurgea, I. et al. RNA Sequencing and Pathway Analysis Identify Important Pathways Involved in Hypertrichosis and Intellectual Disability in Patients with Wiedemann–Steiner Syndrome. Neuromol Med 20, 409–417 (2018). https://doi.org/10.1007/s12017-018-8502-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-018-8502-1