Abstract
Copy number variants (CNVs), involving duplication or deletion of susceptible intervals of the human genome, underlie a range of neurodevelopmental and neuropsychiatric disorders. As accessible in vivo animal models of these disorders often cannot be generated, induced pluripotent stem cell (iPSC) models derived from patients carrying these CNVs can reveal alterations of brain development and neuronal function that contribute to these disorders. CNVs involving deletion versus duplication of a particular genomic interval often result both in distinct clinical phenotypes and in differential phenotypic penetrance. This review initially focuses on CNVs at 15q13.3, which contribute to autism spectrum disorder, attention deficit/hyperactivity disorder, and schizophrenia. Like most CNVs, deletions at 15q13.3 usually cause severe clinical phenotypes, while duplications instead result in highly variable penetrance, with some carriers exhibiting no clinical phenotype. Here, we describe cellular and molecular phenotypes seen in iPSC-derived neuronal models of 15q13.3 duplication and deletion, which may contribute both to the differential clinical consequences and phenotypic penetrance. We then relate this work to many other CNVs involving both duplication and deletion, summarizing findings from iPSC studies and their relationship to clinical phenotype. Together, this work highlights how CNVs involving duplication versus deletion can differentially alter neural development and function to contribute to neuropsychiatric disorders. iPSC-derived neuronal models of these disorders can be used both to understand the underlying neurodevelopmental alterations and to develop pharmacological or molecular approaches for phenotypic rescue that may suggest leads for patient intervention.
Graphical Abstract
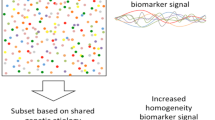
Top: Deletion versus duplication of the same genomic interval results in different clinical phenotypes and degrees of phenotypic penetrance. Example findings schematized. Bottom: iPSC-derived neurons from individuals with these CNVs involving deletion versus duplication likewise often differential phenotypes (increases or decreases) in the categories shown. Figure created with BioRender.com.


Similar content being viewed by others
Data Availability
N/A; no datasets were generated or analyzed during the current study.
Code Availability
N/A
Abbreviations
- ASD :
-
Autism spectrum disorder
- iPSCs :
-
Induced pluripotent stem cells
- CNV :
-
Copy number variant
- CHRNA7 :
-
α-7 Nicotinic acetylcholine receptor subunit
- ID :
-
Intellectual disability
- ADHD :
-
Attention deficit and hyperactivity disorder
- SZ :
-
Schizophrenia
- BD :
-
Bipolar disorder
- EPSC :
-
Excitatory post synaptic current
- IPSC :
-
Inhibitory post synaptic current
- ER :
-
Endoplasmic reticulum
- GABA :
-
Gamma-aminobutyric acid
- NPC :
-
Neural progenitor cell
- UM :
-
Unaffected mother
- AP :
-
Affected proband
- UC-M :
-
Unrelated control-male
- UC-F :
-
Unrelated control-female
References
Anderson, N. C., Chen, P.-F., Meganathan, K., Afshar Saber, W., Petersen, A. J., Bhattacharyya, A., Kroll, K. L., & Sahin, M. (2021). Balancing serendipity and reproducibility: Pluripotent stem cells as experimental systems for intellectual and developmental disorders. Stem Cell Reports, 16(6), 1446–1457. https://doi.org/10.1016/j.stemcr.2021.03.025
Chapman, G., Antony, I., Narasimhan, M., Shen, R., Elman, R., & Kroll, K. L. (2022). Chapter 10: The cellular and molecular neurobiology of autism. In R. Kana (Ed.), The Neuroscience of Autism, (pg (pp. 215–244). Elsevier.
Besag, F. (2017). Epilepsy in patients with autism: Links, risks and treatment challenges. Neuropsychiatric Disease and Treatment, 14, 1–10. https://doi.org/10.2147/NDT.S120509
Bocchi, R., Egervari, K., Carol-Perdiguer, L., Viale, B., Quairiaux, C., De Roo, M., Boitard, M., Oskouie, S., Salmon, P., & Kiss, J. Z. (2017). Perturbed Wnt signaling leads to neuronal migration delay, altered interhemispheric connections and impaired social behavior. Nature Communications, 8(1), 1158. https://doi.org/10.1038/s41467-017-01046-w
Budisteanu, M., Papuc, S. M., Streata, I., Cucu, M., Pirvu, A., Serban-Sosoi, S., Erbescu, A., Andrei, E., Iliescu, C., Ioana, D., Severin, E., Ioana, M., & Arghir, A. (2021). The Phenotypic Spectrum of 15q13.3 Region Duplications: Report of 5 Patients. Genes, 12(7), 1025. https://doi.org/10.3390/genes12071025.
Cavallo, F., Troglio, F., Fagà, G., Fancelli, D., Shyti, R., Trattaro, S., Zanella, M., D’Agostino, G., Hughes, J. M., Cera, M. R., Pasi, M., Gabriele, M., Lazzarin, M., Mihailovich, M., Kooy, F., Rosa, A., Mercurio, C., Varasi, M., & Testa, G. (2020). High-throughput screening identifies histone deacetylase inhibitors that modulate GTF2I expression in 7q11.23 microduplication autism spectrum disorder patient-derived cortical neurons. Molecular Autism, 11(1), 88. https://doi.org/10.1186/s13229-020-00387-6.
Chapman, G., Alsaqati, M., Lunn, S., Singh, T., Linden, S. C., Linden, D. E. J., van den Bree, M. B. M., Ziller, M., Owen, M. J., Hall, J., Harwood, A. J., & Syed, Y. A. (2022). Using induced pluripotent stem cells to investigate human neuronal phenotypes in 1q21.1 deletion and duplication syndrome. Molecular Psychiatry, 27(2), 819–830. https://doi.org/10.1038/s41380-021-01182-2.
Cooper, G. M., Coe, B. P., Girirajan, S., Rosenfeld, J. A., Vu, T. H., Baker, C., Williams, C., Stalker, H., Hamid, R., Hannig, V., Abdel-Hamid, H., Bader, P., McCracken, E., Niyazov, D., Leppig, K., Thiese, H., Hummel, M., Alexander, N., Gorski, J., … Eichler, E. E. (2011). A copy number variation morbidity map of developmental delay. Nature Genetics, 43(9), 838–846. https://doi.org/10.1038/ng.909.
Deshpande, A., Yadav, S., Dao, D. Q., Wu, Z.-Y., Hokanson, K. C., Cahill, M. K., Wiita, A. P., Jan, Y.-N., Ullian, E. M., & Weiss, L. A. (2017). Cellular Phenotypes in Human iPSC-Derived Neurons from a Genetic Model of Autism Spectrum Disorder. Cell Reports, 21(10), 2678–2687. https://doi.org/10.1016/j.celrep.2017.11.037
Drakulic, D., Djurovic, S., Syed, Y. A., Trattaro, S., Caporale, N., Falk, A., Ofir, R., Heine, V. M., Chawner, S. J. R. A., Rodriguez-Moreno, A., van den Bree, M. B. M., Testa, G., Petrakis, S., & Harwood, A. J. (2020). Copy number variants (CNVs): A powerful tool for iPSC-based modelling of ASD. Molecular Autism, 11(1), 42. https://doi.org/10.1186/s13229-020-00343-4
Elitt, M. S., Barbar, L., & Tesar, P. J. (2018). Drug screening for human genetic diseases using iPSC models. Human Molecular Genetics, 27(R2), R89–R98. https://doi.org/10.1093/hmg/ddy186
Fink, J. J., Schreiner, J. D., Bloom, J. E., James, J., Baker, D. S., Robinson, T. M., Lieberman, R., Loew, L. M., Chamberlain, S. J., & Levine, E. S. (2021). Hyperexcitable Phenotypes in Induced Pluripotent Stem Cell-Derived Neurons From Patients With 15q11-q13 Duplication Syndrome, a Genetic Form of Autism. Biological Psychiatry, 90(11), 756–765. https://doi.org/10.1016/j.biopsych.2021.07.018
Frega, M., Linda, K., Keller, J. M., Gümüş-Akay, G., Mossink, B., van Rhijn, J.-R., Negwer, M., Klein Gunnewiek, T., Foreman, K., Kompier, N., Schoenmaker, C., van den Akker, W., van der Werf, I., Oudakker, A., Zhou, H., Kleefstra, T., Schubert, D., van Bokhoven, H., & NadifKasri, N. (2019). Neuronal network dysfunction in a model for Kleefstra syndrome mediated by enhanced NMDAR signaling. Nature Communications, 10(1), 4928. https://doi.org/10.1038/s41467-019-12947-3
Germain, N. D., Chen, P.-F., Plocik, A. M., Glatt-Deeley, H., Brown, J., Fink, J. J., Bolduc, K. A., Robinson, T. M., Levine, E. S., Reiter, L. T., Graveley, B. R., Lalande, M., & Chamberlain, S. J. (2014). Gene expression analysis of human induced pluripotent stem cell-derived neurons carrying copy number variants of chromosome 15q11-q13.1. Molecular Autism, 5(1), 44. https://doi.org/10.1186/2040-2392-5-44.
Gillentine, M. A. (2022). Chapter 13. Exploring 15q13.3 copy number variants in iPSCs. In A. Birbrair (Ed.) Current Topics in iPSCs Technology (pp. 333–360). Elsevier. https://doi.org/10.1016/B978-0-323-99892-5.00017-7
Gillentine, M. A., Berry, L. N., Goin-Kochel, R. P., Ali, M. A., Ge, J., Guffey, D., Rosenfeld, J. A., Hannig, V., Bader, P., Proud, M., Shinawi, M., Graham, B. H., Lin, A., Lalani, S. R., Reynolds, J., Chen, M., Grebe, T., Minard, C. G., Stankiewicz, P., … Schaaf, C. P. (2017). The Cognitive and Behavioral Phenotypes of Individuals with CHRNA7 Duplications. Journal of Autism and Developmental Disorders, 47(3), 549–562. https://doi.org/10.1007/s10803-016-2961-8.
Gillentine, M. A., & Schaaf, C. P. (2015). The human clinical phenotypes of altered CHRNA7 copy number. Biochemical Pharmacology, 97(4), 352–362. https://doi.org/10.1016/j.bcp.2015.06.012
Gillentine, M. A., Yin, J., Bajic, A., Zhang, P., Cummock, S., Kim, J. J., & Schaaf, C. P. (2017). Functional Consequences of CHRNA7 Copy-Number Alterations in Induced Pluripotent Stem Cells and Neural Progenitor Cells. The American Journal of Human Genetics, 101(6), 874–887. https://doi.org/10.1016/j.ajhg.2017.09.024
Gregoric Kumperscak H., Krgovic D., Drobnic Radobuljac M., Senica N., Zagorac A., Kokalj Vokac N. (2021) CNVs and Chromosomal Aneuploidy in Patients With Early-Onset Schizophrenia and Bipolar Disorder: Genotype-Phenotype Associations. Frontiers in Psychiatry, 11, pg. 606372. https://doi.org/10.3389/fpsyt.2020.606372
Hakamata, Y., Nakai, J., Takeshima, H., & Imoto, K. (1992). Primary structure and distribution of a novel ryanodine receptor/calcium release channel from rabbit brain. FEBS Letters, 312(2–3), 229–235. https://doi.org/10.1016/0014-5793(92)80941-9
Hassfurther, A., Komini, E., Fischer, J., & Leipoldt, M. (2015). Clinical and Genetic Heterogeneity of the 15q13.3 Microdeletion Syndrome. Molecular Syndromology, 6(5), 222–228. https://doi.org/10.1159/000443343.
Jang, Y.-Y., & Ye, Z. (2016). Gene correction in patient-specific iPSCs for therapy development and disease modeling. Human Genetics, 135(9), 1041–1058. https://doi.org/10.1007/s00439-016-1691-5
Kaminsky, E. B., Kaul, V., Paschall, J., Church, D. M., Bunke, B., Kunig, D., Moreno-De-Luca, D., Moreno-De-Luca, A., Mulle, J. G., Warren, S. T., Richard, G., Compton, J. G., Fuller, A. E., Gliem, T. J., Huang, S., Collinson, M. N., Beal, S. J., Ackley, T., Pickering, D. L., … Martin, C. L. (2011). An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in Medicine, 13(9), 777–784. https://doi.org/10.1097/GIM.0b013e31822c79f9.
Kathuria, A., Nowosiad, P., Jagasia, R., Aigner, S., Taylor, R. D., Andreae, L. C., Gatford, N. J. F., Lucchesi, W., Srivastava, D. P., & Price, J. (2018). Stem cell-derived neurons from autistic individuals with SHANK3 mutation show morphogenetic abnormalities during early development. Molecular Psychiatry, 23(3), 735–746. https://doi.org/10.1038/mp.2017.185
Khan, T. A., Revah, O., Gordon, A., Yoon, S.-J., Krawisz, A. K., Goold, C., Sun, Y., Kim, C. H., Tian, Y., Li, M.-Y., Schaepe, J. M., Ikeda, K., Amin, N. D., Sakai, N., Yazawa, M., Kushan, L., Nishino, S., Porteus, M. H., Rapoport, J. L., … Paşca, S. P. (2020). Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nature Medicine, 26(12), 1888–1898. https://doi.org/10.1038/s41591-020-1043-9.
Khattak, S., Brimble, E., Zhang, W., Zaslavsky, K., Strong, E., Ross, P. J., Hendry, J., Mital, S., Salter, M. W., Osborne, L. R., & Ellis, J. (2015). Human induced pluripotent stem cell derived neurons as a model for Williams-Beuren syndrome. Molecular Brain, 8(1), 77. https://doi.org/10.1186/s13041-015-0168-0
Köhler, S., Gargano, M., Matentzoglu, N., Carmody, L. C., Lewis-Smith, D., Vasilevsky, N. A., Danis, D., Balagura, G., Baynam, G., Brower, A. M., Callahan, T. J., Chute, C. G., Est, J. L., Galer, P. D., Ganesan, S., Griese, M., Haimel, M., Pazmandi, J., Hanauer, M., … Robinson, P. N. (2021). The Human Phenotype Ontology in 2021. Nucleic Acids Research, 49(D1), D1207–D1217. https://doi.org/10.1093/nar/gkaa1043.
Krebs, J., Agellon, L. B., & Michalak, M. (2015). Ca2+ homeostasis and endoplasmic reticulum (ER) stress: An integrated view of calcium signaling. Biochemical and Biophysical Research Communications, 460(1), 114–121. https://doi.org/10.1016/j.bbrc.2015.02.004
Kushima, I., Aleksic, B., Nakatochi, M., Shimamura, T., Okada, T., Uno, Y., Morikawa, M., Ishizuka, K., Shiino, T., Kimura, H., Arioka, Y., Yoshimi, A., Takasaki, Y., Yu, Y., Nakamura, Y., Yamamoto, M., Iidaka, T., Iritani, S., Inada, T., … Ozaki, N. (2018). Comparative Analyses of Copy-Number Variation in Autism Spectrum Disorder and Schizophrenia Reveal Etiological Overlap and Biological Insights. Cell Reports, 24(11), 2838–2856. https://doi.org/10.1016/j.celrep.2018.08.022.
Lewis, E. M. A., Meganathan, K., Baldridge, D., Gontarz, P., Zhang, B., Bonni, A., Constantino, J. N., & Kroll, K. L. (2019). Cellular and molecular characterization of multiplex autism in human induced pluripotent stem cell-derived neurons. Molecular Autism, 10(1), 51. https://doi.org/10.1186/s13229-019-0306-0
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., & Zhao, X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduction and Targeted Therapy, 5(1), 1. https://doi.org/10.1038/s41392-019-0089-y
Loh, Y.-H., Agarwal, S., Park, I.-H., Urbach, A., Huo, H., Heffner, G. C., Kim, K., Miller, J. D., Ng, K., & Daley, G. Q. (2009). Generation of induced pluripotent stem cells from human blood. Blood, 113(22), 5476–5479. https://doi.org/10.1182/blood-2009-02-204800
Lowther, C., Costain, G., Stavropoulos, D. J., Melvin, R., Silversides, C. K., Andrade, D. M., So, J., Faghfoury, H., Lionel, A. C., Marshall, C. R., Scherer, S. W., & Bassett, A. S. (2015). Delineating the 15q13.3 microdeletion phenotype: A case series and comprehensive review of the literature. Genetics in Medicine, 17(2), 149–157. https://doi.org/10.1038/gim.2014.83.
Malhotra, D., & Sebat, J. (2012). CNVs: Harbingers of a rare variant revolution in psychiatric genetics. Cell, 148(6), 1223–1241. https://doi.org/10.1016/j.cell.2012.02.039
Malwade, S., Gasthaus, J., Bellardita, C., Andelic, M., Moric, B., Korshunova, I., Kiehn, O., Vasistha, N. A., & Khodosevich, K. (2022). Identification of Vulnerable Interneuron Subtypes in 15q13.3 Microdeletion Syndrome Using Single-Cell Transcriptomics. Biological Psychiatry, 91(8), 727–739. https://doi.org/10.1016/j.biopsych.2021.09.012.
Meganathan, K., Prakasam, R., Baldridge, D., Gontarz, P., Zhang, B., Urano, F., Bonni, A., Maloney, S. E., Turner, T. N., Huettner, J. E., Constantino, J. N., & Kroll, K. L. (2021). Altered neuronal physiology, development, and function associated with a common chromosome 15 duplication involving CHRNA7. BMC Biology, 19(1), 147. https://doi.org/10.1186/s12915-021-01080-7
Parenti, I., Rabaneda, L. G., Schoen, H., & Novarino, G. (2020). Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends in Neurosciences, 43(8), 608–621. https://doi.org/10.1016/j.tins.2020.05.004
Pös, O., Radvanszky, J., Buglyó, G., Pös, Z., Rusnakova, D., Nagy, B., & Szemes, T. (2021). DNA copy number variation: Main characteristics, evolutionary significance, and pathological aspects. Biomedical Journal, 44(5), 548–559. https://doi.org/10.1016/j.bj.2021.02.003
Rein, B., & Yan, Z. (2020). 16p11.2 Copy Number Variations and Neurodevelopmental Disorders. Trends in Neurosciences, 43(11), 886–901. https://doi.org/10.1016/j.tins.2020.09.001.
Rosenfeld, J. A., Coe, B. P., Eichler, E. E., Cuckle, H., & Shaffer, L. G. (2013). Estimates of penetrance for recurrent pathogenic copy-number variations. Genetics in Medicine, 15(6), 478–481. https://doi.org/10.1038/gim.2012.164
Sabitha, K. R., Shetty, A. K., & Upadhya, D. (2021). Patient-derived iPSC modeling of rare neurodevelopmental disorders: Molecular pathophysiology and prospective therapies. Neuroscience & Biobehavioral Reviews, 121, 201–219. https://doi.org/10.1016/j.neubiorev.2020.12.025
Shcheglovitov, A., Shcheglovitova, O., Yazawa, M., Portmann, T., Shu, R., Sebastiano, V., Krawisz, A., Froehlich, W., Bernstein, J. A., Hallmayer, J. F., & Dolmetsch, R. E. (2013). SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature, 503(7475), 267–271. https://doi.org/10.1038/nature12618
Smajlagić, D., Lavrichenko, K., Berland, S., Helgeland, Ø., Knudsen, G. P., Vaudel, M., Haavik, J., Knappskog, P. M., Njølstad, P. R., Houge, G., & Johansson, S. (2021). Population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders in 12,252 newborns and their parents. European Journal of Human Genetics, 29(1), 205–215. https://doi.org/10.1038/s41431-020-00707-7
Sui, Y., & Peng, S. (2021). A Mechanism Leading to Changes in Copy Number Variations Affected by Transcriptional Level Might Be Involved in Evolution, Embryonic Development, Senescence, and Oncogenesis Mediated by Retrotransposons. Frontiers in Cell and Developmental Biology, 9, 618113. https://doi.org/10.3389/fcell.2021.618113
Sundberg, M., Pinson, H., Smith, R. S., Winden, K. D., Venugopal, P., Tai, D. J. C., Gusella, J. F., Talkowski, M. E., Walsh, C. A., Tegmark, M., & Sahin, M. (2021). 16p11.2 deletion is associated with hyperactivation of human iPSC-derived dopaminergic neuron networks and is rescued by RHOA inhibition in vitro. Nature Communications, 12(1), 2897. https://doi.org/10.1038/s41467-021-23113-z.
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., & Yamanaka, S. (2007). Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell, 131(5), 861–872. https://doi.org/10.1016/j.cell.2007.11.019
Takumi, T., & Tamada, K. (2018). CNV biology in neurodevelopmental disorders. Current Opinion in Neurobiology, 48, 183–192. https://doi.org/10.1016/j.conb.2017.12.004
Turkalj, L., Mehta, M., Matteson, P., Prem, S., Williams, M., Connacher, R. J., DiCicco-Bloom, E., & Millonig, J. H. (2020). Using iPSC-Based Models to Understand the Signaling and Cellular Phenotypes in Idiopathic Autism and 16p11.2 Derived Neurons. In E. DiCicco-Bloom & J. H. Millonig (Eds.), Neurodevelopmental Disorders (Vol. 25, pp. 79–107). Springer International Publishing. https://doi.org/10.1007/978-3-030-45493-7_4.
Wilfert, A. B., Sulovari, A., Turner, T. N., Coe, B. P., & Eichler, E. E. (2017). Recurrent de novo mutations in neurodevelopmental disorders: Properties and clinical implications. Genome Medicine, 9, 101. https://doi.org/10.1186/s13073-017-0498-x
Zhang, F., & Lupski, J. R. (2015). Non-coding genetic variants in human disease: Figure 1. Human Molecular Genetics, 24(R1), R102–R110. https://doi.org/10.1093/hmg/ddv259
Zhang, S., Zhang, X., Purmann, C., Ma, S., Shrestha, A., Davis, K. N., Ho, M., Huang, Y., Pattni, R., Wong, W. H., Bernstein, J. A., Hallmayer, J., & Urban, A. E. (2021). Network Effects of the 15q13.3 Microdeletion on the Transcriptome and Epigenome in Human-Induced Neurons. Biological Psychiatry, 89(5), 497–509. https://doi.org/10.1016/j.biopsych.2020.06.021.
Zhu, P., Guo, H., Ren, Y., Hou, Y., Dong, J., Li, R., Lian, Y., Fan, X., Hu, B., Gao, Y., Wang, X., Wei, Y., Liu, P., Yan, J., Ren, X., Yuan, P., Yuan, Y., Yan, Z., Wen, L., … Tang, F. (2018). Single-cell DNA methylome sequencing of human preimplantation embryos. Nature Genetics, 50(1), 12–19. https://doi.org/10.1038/s41588-017-0007-6.
Ziats, M. N., Goin-Kochel, R. P., Berry, L. N., Ali, M., Ge, J., Guffey, D., Rosenfeld, J. A., Bader, P., Gambello, M. J., Wolf, V., Penney, L. S., Miller, R., Lebel, R. R., Kane, J., Bachman, K., Troxell, R., Clark, G., Minard, C. G., Stankiewicz, P., … Schaaf, C. P. (2016). The complex behavioral phenotype of 15q13.3 microdeletion syndrome. Genetics in Medicine, 18(11), 1111–1118. https://doi.org/10.1038/gim.2016.9.
Funding
This work was supported by NIH grants R01NS114551 and R01MH124808 (KLK), and by NIH P50HD103525 to John N Constantino and Christina Gurnett (KLK serves as a project PI for the Model Systems Core).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design, IA and GC outlined the manuscript, IA, GC, RP, and MN gathered the literature and sources and IA, MN, RS, RP, KK, and KLK drafted and edited the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics Approval
Not applicable (N/A)
Consent to Participate
N/A
Consent for Publication
N/A
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Irene Antony, Mishka Narasimhan, Renata Shen, Ramachandran Prakasam these authors contributed equally to this work.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Antony, I., Narasimhan, M., Shen, R. et al. Duplication Versus Deletion Through the Lens of 15q13.3: Clinical and Research Implications of Studying Copy Number Variants Associated with Neuropsychiatric Disorders in Induced Pluripotent Stem Cell-Derived Neurons. Stem Cell Rev and Rep 19, 639–650 (2023). https://doi.org/10.1007/s12015-022-10475-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-022-10475-0