Abstract
Epigenetic regulation of gene expression represents an important mechanism in the maintenance of stem cell function. Alterations in epigenetic regulation contribute to the pathogenesis of hematological malignancies. Plant homeodomain finger protein 6 (PHF6) is a member of the plant homeodomain (PHD)-like zinc finger family of proteins that is involved in transcriptional regulation through the modification of the chromatin state. Germline mutation of PHF6 is the causative genetic alteration of the X-linked mental retardation Borjeson–Forssman–Lehmann syndrome (BFLS). Somatic mutations in PHF6 are identified in human leukemia, such as adult T-cell acute lymphoblastic leukemia (T-ALL, ~ 38%), pediatric T-ALL (~ 16%), acute myeloid leukemia (AML, ~ 3%), chronic myeloid leukemia (CML, ~ 2.5%), mixed phenotype acute leukemia (MPAL, ~ 20%), and high-grade B-cell lymphoma (HGBCL, ~ 3%). More recent studies imply an oncogenic effect of PHF6 in B-cell acute lymphoblastic leukemia (B-ALL) and solid tumors. These data demonstrate that PHF6 could act as a double-edged sword, either a tumor suppressor or an oncogene, in a lineage-dependent manner. However, the underlying mechanisms of PHF6 in normal hematopoiesis and leukemogenesis remain largely unknown. In this review, we summarize current knowledge of PHF6, emphasizing the role of PHF6 in hematological malignancies.
Graphical abstract
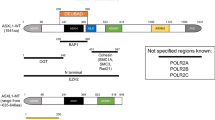
Epigenetic regulation of PHF6 in B-ALL. PHF6 maintains a chromatin structure that is permissive to B-cell identity genes, but not T-cell-specific genes (left). Loss of PHF6 leads to aberrant expression of B-cell- and T-cell-specific genes resulting from lineage promiscuity and binding of T-cell transcription factors (right).



Similar content being viewed by others
Data Availability
Not applicable.
References
Lower, K. M., Turner, G., Kerr, B. A., et al. (2002). Mutations in PHF6 are associated with Börjeson-Forssman-Lehmann syndrome. Nature Genetics, 32(4), 661–665. https://doi.org/10.1038/ng1040
Vallée, D., Chevrier, E., Graham, G. E., et al. (2004). A novel PHF6 mutation results in enhanced exon skipping and mild Börjeson-Forssman-Lehmann syndrome. Journal of Medical Genetics, 41(10), 778–783. https://doi.org/10.1136/jmg.2004.020370
Voss, A. K., Gamble, R., Collin, C., et al. (2007). Protein and gene expression analysis of Phf6, the gene mutated in the Börjeson-Forssman-Lehmann Syndrome of intellectual disability and obesity. Gene Expression Patterns, 7(8), 858–871. https://doi.org/10.1016/j.modgep.2007.06.007
Todd, M. A., & Picketts, D. J. (2012). PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. Journal of Proteome Research, 11(8), 4326–4337. https://doi.org/10.1021/pr3004369
Perry, J. (2006). The Epc-N domain: A predicted protein-protein interaction domain found in select chromatin associated proteins. BMC Genomics, 7, 6. https://doi.org/10.1186/1471-2164-7-6
Kurzer, J. H., & Weinberg, O. K. (2021). PHF6 Mutations in Hematologic Malignancies. Frontiers in Oncology, 11, 704471. https://doi.org/10.3389/fonc.2021.704471
Grossmann, V., Haferlach, C., Weissmann, S., et al. (2013). The molecular profile of adult T-cell acute lymphoblastic leukemia: Mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes, Chromosomes & Cancer, 52(4), 410–422. https://doi.org/10.1002/gcc.22039
Van Vlierberghe, P., Palomero, T., Khiabanian, H., et al. (2010). PHF6 mutations in T-cell acute lymphoblastic leukemia. Nature Genetics, 42(4), 338–342. https://doi.org/10.1038/ng.542
Loontiens, S., Dolens, A. C., Strubbe, S., et al. (2020). PHF6 Expression Levels Impact Human Hematopoietic Stem Cell Differentiation. Frontiers in Cellular Developmental Biology, 8, 599472. https://doi.org/10.3389/fcell.2020.599472
Visootsak, J., Rosner, B., Dykens, E., et al. (2004). Clinical and behavioral features of patients with Borjeson-Forssman-Lehmann syndrome with mutations in PHF6. Journal of Pediatrics, 145(6), 819–825. https://doi.org/10.1016/j.jpeds.2004.07.041
Soto-Feliciano, Y. M., Bartlebaugh, J. M. E., Liu, Y., et al. (2017). PHF6 regulates phenotypic plasticity through chromatin organization within lineage-specific genes. Genes & Development, 31(10), 973–989. https://doi.org/10.1101/gad.295857.117
Liu, Z., Li, F., Ruan, K., et al. (2014). Structural and functional insights into the human Börjeson-Forssman-Lehmann syndrome-associated protein PHF6. Journal of Biological Chemistry, 289(14), 10069–10083. https://doi.org/10.1074/jbc.M113.535351
Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Côté, J., & Wang, W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Molecular Cell, 2(6), 851–861. https://doi.org/10.1016/s1097-2765(00)80299-3
Potts, R. C., Zhang, P., Wurster, A. L., et al. (2011). CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS One, 6(9), e24515. https://doi.org/10.1371/journal.pone.0024515
Yoshida, T., Hazan, I., Zhang, J., et al. (2008). The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes & Development, 22(9), 1174–1189. https://doi.org/10.1101/gad.1642808
Shi, X., Hong, T., Walter, K. L., et al. (2006). ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature, 442(7098), 96–99. https://doi.org/10.1038/nature04835
Oh, S., Boo, K., Kim, J., et al. (2020). The chromatin-binding protein PHF6 functions as an E3 ubiquitin ligase of H2BK120 via H2BK12Ac recognition for activation of trophectodermal genes. Nucleic Acids Research, 48(16), 9037–9052. https://doi.org/10.1093/nar/gkaa626
Wang, J., Leung, J. W., Gong, Z., Feng, L., Shi, X., & Chen, J. (2013). PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. Journal of Biological Chemistry, 288(5), 3174–3183. https://doi.org/10.1074/jbc.M112.414839
Zhang, C., Mejia, L. A., Huang, J., et al. (2013). The X-linked intellectual disability protein PHF6 associates with the PAF1 complex and regulates neuronal migration in the mammalian brain. Neuron, 78(6), 986–993. https://doi.org/10.1016/j.neuron.2013.04.021
Warmerdam, D. O., Alonso-de Vega, I., Wiegant, W. W., et al. (2020). PHF6 promotes non-homologous end joining and G2 checkpoint recovery. EMBO Reports, 21(1), e48460. https://doi.org/10.15252/embr.201948460
Turner, G., Lower, K. M., White, S. M., et al. (2004). The clinical picture of the Börjeson-Forssman-Lehmann syndrome in males and heterozygous females with PHF6 mutations. Clinical Genetics, 65(3), 226–232. https://doi.org/10.1111/j.0009-9163.2004.00215.x
Borjeson, M., Forssman, H., & Lehmann, O. (1962). An X-linked, recessively inherited syndrome characterized by grave mental deficiency, epilepsy, and endocrine disorder. Acta Medica Scandinavica, 171, 13–21. https://doi.org/10.1111/j.0954-6820.1962.tb04162.x
Gécz, J., Turner, G., Nelson, J., & Partington, M. (2006). The Börjeson-Forssman-Lehman syndrome (BFLS, MIM #301900). European Journal of Human Genetics, 14(12), 1233–1237. https://doi.org/10.1038/sj.ejhg.5201639
Todd, M. A., Ivanochko, D., & Picketts, D. J. (2015). PHF6 Degrees of Separation: The Multifaceted Roles of a Chromatin Adaptor Protein. Genes (Basel), 6(2), 325–352. https://doi.org/10.3390/genes6020325
Berland, S. (2011). PHF6 deletions may cause borjeson-forssman-lehmann syndrome in females. Molecular Syndromology. https://doi.org/10.1159/000330111
Mt, C. (2009). Further clinical delineation of the Börjeson–Forssman–Lehmann syndrome in patients with PHF6 mutations. American Journal of Medical Genetics. https://doi.org/10.1002/ajmg.a.32624
Crawford, J., Lower, K. M., Hennekam, R. C., et al. (2006). Mutation screening in Borjeson-Forssman-Lehmann syndrome: Identification of a novel de novo PHF6 mutation in a female patient. Journal of Medical Genetics, 43(3), 238–243. https://doi.org/10.1136/jmg.2005.033084
Zweier, C., Kraus, C., Brueton, L., et al. (2013). A new face of Borjeson-Forssman-Lehmann syndrome? De novo mutations in PHF6 in seven females with a distinct phenotype. Journal of Medical Genetics, 50(12), 838–847. https://doi.org/10.1136/jmedgenet-2013-101918
Cheng, C., Deng, P. Y., Ikeuchi, Y., et al. (2018). Characterization of a Mouse Model of Börjeson-Forssman-Lehmann Syndrome. Cell Reports, 25(6), 1404-1414.e6. https://doi.org/10.1016/j.celrep.2018.10.043
McRae, H. M., Eccles, S., Whitehead, L., et al. (2020). Downregulation of the GHRH/GH/IGF1 axis in a mouse model of Börjeson-Forssman-Lehman syndrome. Development, 147(21). https://doi.org/10.1242/dev.187021
Ferrando, A. A., Neuberg, D. S., Staunton, J., et al. (2002). Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell, 1(1), 75–87. https://doi.org/10.1016/s1535-6108(02)00018-1
Wendorff, A. A., Quinn, S. A., Rashkovan, M., et al. (2019). Phf6 Loss Enhances HSC Self-Renewal Driving Tumor Initiation and Leukemia Stem Cell Activity in T-ALL. Cancer Discovery, 9(3), 436–451. https://doi.org/10.1158/2159-8290.Cd-18-1005
Ahmed, R., Sarwar, S., Hu, J., et al. (2021). Transgenic mice with an R342X mutation in Phf6 display clinical features of Börjeson-Forssman-Lehmann Syndrome. Human Molecular Genetics, 30(7), 575–594. https://doi.org/10.1093/hmg/ddab081
Ogilvy, S., Elefanty, A. G., Visvader, J., Bath, M. L., Harris, A. W., & Adams, J. M. (1998). Transcriptional regulation of vav, a gene expressed throughout the hematopoietic compartment. Blood, 91(2), 419–430.
Van Vlierberghe, P., Patel, J., Abdel-Wahab, O., et al. (2011). PHF6 mutations in adult acute myeloid leukemia. Leukemia, 25(1), 130–134. https://doi.org/10.1038/leu.2010.247
Karrman, K., Castor, A., Behrendtz, M., et al. (2015). Deep sequencing and SNP array analyses of pediatric T-cell acute lymphoblastic leukemia reveal NOTCH1 mutations in minor subclones and a high incidence of uniparental isodisomies affecting CDKN2A. Journal of Hematology & Oncology, 8, 42. https://doi.org/10.1186/s13045-015-0138-0
Yoo, N. J., Kim, Y. R., & Lee, S. H. (2012). Somatic mutation of PHF6 gene in T-cell acute lymphoblatic leukemia, acute myelogenous leukemia and hepatocellular carcinoma. Acta Oncologica, 51(1), 107–111. https://doi.org/10.3109/0284186x.2011.592148
Spinella, J. F., Cassart, P., Richer, C., et al. (2016). Genomic characterization of pediatric T-cell acute lymphoblastic leukemia reveals novel recurrent driver mutations. Oncotarget, 7(40), 65485–65503. https://doi.org/10.18632/oncotarget.11796
Wang, Q., Qiu, H., Jiang, H., et al. (2011). Mutations of PHF6 are associated with mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214 in T-cell acute lymphoblastic leukemia. Haematologica, 96(12), 1808–1814. https://doi.org/10.3324/haematol.2011.043083
Alexander, T. B., Gu, Z., Iacobucci, I., et al. (2018). The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature, 562(7727), 373–379. https://doi.org/10.1038/s41586-018-0436-0
Xiao, W., Bharadwaj, M., Levine, M., et al. (2018). PHF6 and DNMT3A mutations are enriched in distinct subgroups of mixed phenotype acute leukemia with T-lineage differentiation. Blood Advances, 2(23), 3526–3539. https://doi.org/10.1182/bloodadvances.2018023531
Mi, X., Griffin, G., Lee, W., et al. (2018). Genomic and clinical characterization of B/T mixed phenotype acute leukemia reveals recurrent features and T-ALL like mutations. American Journal of Hematology, 93(11), 1358–1367. https://doi.org/10.1002/ajh.25256
Mori, T., Nagata, Y., Makishima, H., et al. (2016). Somatic PHF6 mutations in 1760 cases with various myeloid neoplasms. Leukemia, 30(11), 2270–2273. https://doi.org/10.1038/leu.2016.212
Xiao, W., Pastore, F., Getta, B., et al. (2017). PHF6 Mutations Defines a Subgroup of Mixed Phenotype of Acute Leukemia with Aberrant T-Cell Differentiation. Blood, 130(Supplement 1), 1384–1384. https://doi.org/10.1182/blood.V130.Suppl_1.1384.1384
de Rooij, J. D., van den Heuvel-Eibrink, M. M., van de Rijdt, N. K., et al. (2016). PHF6 mutations in paediatric acute myeloid leukaemia. British Journal of Haematology, 175(5), 967–971. https://doi.org/10.1111/bjh.13891
Li, X., Yao, H., Chen, Z., Wang, Q., Zhao, Y., & Chen, S. (2013). Somatic mutations of PHF6 in patients with chronic myeloid leukemia in blast crisis. Leukaemia & Lymphoma, 54(3), 671–672. https://doi.org/10.3109/10428194.2012.725203
Huh, H. J., Lee, S. H., Yoo, K. H., et al. (2013). Gene mutation profiles and prognostic implications in Korean patients with T-lymphoblastic leukemia. Annals of Hematology, 92(5), 635–644. https://doi.org/10.1007/s00277-012-1664-2
Stengel, A., Kern, W., Meggendorfer, M., Haferlach, T., & Haferlach, C. (2017). High Grade B Cell Lymphoma with MYC and BCL2 and/or BCL6 Rearrangements Depict a High Complexity on the Cytogenetic, but Not on the Molecular Genetic Level and Show MYC Mutations As Prognostic Marker. Blood, 130(Supplement 1), 4001–4001. https://doi.org/10.1182/blood.V130.Suppl_1.4001.4001
Ueno, H., Yoshida, K., Shiozawa, Y., et al. (2020). Landscape of driver mutations and their clinical impacts in pediatric B-cell precursor acute lymphoblastic leukemia. Blood Advances, 4(20), 5165–5173. https://doi.org/10.1182/bloodadvances.2019001307
Weinberg, O. K., & Arber, D. A. (2010). Mixed-phenotype acute leukemia: Historical overview and a new definition. Leukemia, 24(11), 1844–1851. https://doi.org/10.1038/leu.2010.202
Eckstein, O. S., Wang, L., Punia, J. N., et al. (2016). Mixed-phenotype acute leukemia (MPAL) exhibits frequent mutations in DNMT3A and activated signaling genes. Experimental Hematology, 44(8), 740–744. https://doi.org/10.1016/j.exphem.2016.05.003
De Kouchkovsky, I., & Abdul-Hay, M. (2016). Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer Journal, 6(7), e441. https://doi.org/10.1038/bcj.2016.50
Chereda, B., & Melo, J. V. (2015). Natural course and biology of CML. Annals of Hematology, 94(Suppl 2), S107–S121. https://doi.org/10.1007/s00277-015-2325-z
Yu, Q., Yin, L., Jian, Y., Li, P., Zeng, W., & Zhou, J. (2019). Downregulation of PHF6 Inhibits Cell Proliferation and Migration in Hepatocellular Carcinoma. Cancer Biotherapy & Radiopharmaceuticals, 34(4), 245–251. https://doi.org/10.1089/cbr.2018.2671
Hajjari, M. (2015). The potential role of PHF6 as an oncogene: A genotranscriptomic/proteomic meta-analysis. Tumor Biology. https://doi.org/10.1007/s13277-015-4250-0
Patel, J. P., Gönen, M., Figueroa, M. E., et al. (2012). Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. New England Journal of Medicine, 366(12), 1079–1089. https://doi.org/10.1056/NEJMoa1112304
Xiang, J., Wang, G., Xia, T., & Chen, Z. (2019). The depletion of PHF6 decreases the drug sensitivity of T-cell acute lymphoblastic leukemia to prednisolone. Biomedicine & Pharmacotherapy, 109, 2210–2217. https://doi.org/10.1016/j.biopha.2018.11.083
Meacham, C. E., Lawton, L. N., Soto-Feliciano, Y. M., et al. (2015). A genome-scale in vivo loss-of-function screen identifies Phf6 as a lineage-specific regulator of leukemia cell growth. Genes & Development, 29(5), 483–488. https://doi.org/10.1101/gad.254151.114
Hsu, Y. C., Chen, T. C., Lin, C. C., et al. (2019). Phf6-null hematopoietic stem cells have enhanced self-renewal capacity and oncogenic potentials. Blood Advances, 3(15), 2355–2367. https://doi.org/10.1182/bloodadvances.2019000391
McRae, H. M., Garnham, A. L., Hu, Y., et al. (2019). PHF6 regulates hematopoietic stem and progenitor cells and its loss synergizes with expression of TLX3 to cause leukemia. Blood, 133(16), 1729–1741. https://doi.org/10.1182/blood-2018-07-860726
Miyagi, S., Sroczynska, P., Kato, Y., et al. (2019). The chromatin-binding protein Phf6 restricts the self-renewal of hematopoietic stem cells. Blood, 133(23), 2495–2506. https://doi.org/10.1182/blood.2019000468
Funding
This work was supported by grants from the National Institutes of Health (HL149318, HL158081, and CA172408 to F.-C.Y.).
Author information
Authors and Affiliations
Contributions
Yusra A. Eisa, Ying Guo and Feng-Chun Yang wrote the manuscript and gave final approval of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Institutional Review Board
Not applicable.
Informed Consent
Not applicable.
Conflicts of Interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Eisa, Y.A., Guo, Y. & Yang, FC. The Role of PHF6 in Hematopoiesis and Hematologic Malignancies. Stem Cell Rev and Rep 19, 67–75 (2023). https://doi.org/10.1007/s12015-022-10447-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-022-10447-4