Abstract
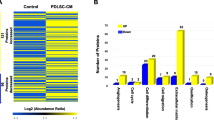
There is substantial evidence supporting the anti-inflammatory and regenerative potential of dental pulp stem cells (DPSCs) through direct cell transplantation or paracrine action. However, DPSC secretome profile remains inadequately studied. This study provides proteomic profiling of the human DPSC secretome by comparatively analysising cell lysates and respective culture supernatants (i.e. conditioned media-CM) under variable oxygen tension conditions (normoxia-20% O2/CM_Norm vs. hypoxia 2% O2/CM_Hyp) and/or stimulation with Tumor Necrosis Factor alpha (TNF-α). DPSC-CM samples and respective crude lysates (DPSC-CL) were collected and subjected to SDS-PAGE, followed by LC-MS/MS analysis. The identified proteins were analyzed by Gene Ontology, Reactome, and String databases. The anti-inflammatory properties of DPSC-CMs were validated via an in vitro RAW_246.7 murine macrophages model through evaluation of the expression of pro-and anti-inflammatory markers by real-time PCR. Results showed a total of 2413 proteins identified in CM_Norm, 2479 in CM_Norm+TNF-α, 1642 in CM_Hyp, and 2002 in CM_Hyp + TNF-α samples. CM_Norm contained 122 proteins statistically significantly upregulated compared to the CM_Hyp and involved in pathways related to “ECM organization”, “cellular response to hypoxia”, and “IL signaling”. Functional network analysis showed that TGFβ1, TIMP1 and TIMP2 were key nodes among proteins significantly upregulated in the CM_Norm compared to the CM_Hyp, interacting with more than 10 proteins, each. DPSC-CM application in the in vitro RAW_246.7 model decreased the expression of pro-inflammatory markers (MMP-3, MMP-9, MMP-13, MCP-1), while increasing anti-inflammatory markers (IL-10). Overall, DPSC-CM collected under normoxic conditions is enriched with anti-inflammatory, tissue repair and regenerative factors, which prompts further investigation on its therapeutic applications.
Graphical abstract









Similar content being viewed by others
Data Availability
Data can be made available from the corresponding author upon reasonable request.
References
Caplan, A. I. (1991). Mesenchymal stem cells. Journal of Orthopaedic Research, 9, 641–650. https://doi.org/10.1016/B978-0-12-374729-7.00029-9
https://clinicaltrials.gov/ct2/results?cond=&term=Mesenchymal+Stem+Cells&cntry=&state=&city=&dist=. (n.d.).
Caplan, A. I. (2010). What’s in a name? Tissue Engineering Parts A, 16(8), 2415–2417. https://doi.org/10.1089/ten.tea.2010.0216
Murry, C. E., Soonpaa, M. H., Reinecke, H., Nakajima, H., Nakajima, H. O., Rubart, M., Pasumarthi, K. B. S., Ismail Virag, J., Bartelmez, S. H., Poppa, V., Bradford, G., Dowell, J. D., Williams, D. A., & Field, L. J. (2004). Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature, 428(6983), 664–668. https://doi.org/10.1038/nature02446
Gnecchi, M., He, H., Liang, O. D., Melo, L. G., Morello, F., Mu, H., et al. (2005). Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nature Medicine, 11, 367–368. https://doi.org/10.1096/fj.04-2702fje
Gnecchi, M., He, H., Noiseux, N., Liang, O. D., Zhang, L., Morello, F., Mu, H., Melo, L. G., Pratt, R. E., Ingwall, J. S., & Dzau, V. J. (2006). Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. The FASEB Journal, 20(6), 661–669. https://doi.org/10.1096/fj.05-5211com
Fabre, T., Kared, H., Friedman, S. L., & Shoukry, N. H. (2014). IL-17A enhances the expression of pro-fibrotic genes through upregulation of the TGF-β receptor on hepatic stellate cells in a JNK-dependent manner. Journal of Immunology, 193(8), 3925–3933. https://doi.org/10.1038/jid.2014.371
Fabre, T., Molina, M. F., Soucy, G., Goulet, J. P., Willems, B., Villeneuve, J. P., Bilodeau, M., & Shoukry, N. H. (2018). Type 3 cytokines IL-17A and IL-22 drive TGF-–dependent liver fibrosis. Science Immunology, 3(28), 1–16. https://doi.org/10.1126/sciimmunol.aar7754
Xin, H., Li, Y., Cui, Y., Yang, J. J., Zhang, Z. G., & Chopp, M. (2013). Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. Journal of Cerebral Blood Flow and Metabolism, 33(11), 1711–1715. https://doi.org/10.1038/jcbfm.2013.152
Cruz, F. F., Borg, Z. D., Goodwin, M., Sokocevic, D., Wagner, D. E., Coffey, A., Antunes, M., Robinson, K. L., Mitsialis, S. A., Kourembanas, S., Thane, K., Hoffman, A. M., McKenna, D. H., Rocco, P. R. M., & Weiss, D. J. (2015). Systemic Administration of Human Bone Marrow-Derived Mesenchymal Stromal Cell Extracellular Vesicles Ameliorates Aspergillus Hyphal Extract-Induced Allergic Airway Inflammation in immunocompetent mice. Stem Cells Translational Medicine, 4(11), 1302–1316. https://doi.org/10.5966/sctm.2014-0280
Pattappa, G., Johnstone, B., Zellner, J., Docheva, D., & Angele, P. (2019). The importance of physioxia in mesenchymal stem cell chondrogenesis and the mechanisms controlling its response. International Journal of Molecular Sciences, 20(3), 1–28. https://doi.org/10.3390/ijms20030484
Lotfinia, M., Lak, S., Ghahhari, N. M., Johari, B., Maghsood, F., Parsania, S., … Kadivar, M. (2017). Hypoxia pre-conditioned embryonic mesenchymal stem cell secretome reduces IL-10 production by peripheral blood mononuclear cells. Iranian Biomedical Journal, 21(1), 24–31. https://doi.org/10.18869/acadpub.ibj.21.1.24.
Bakopoulou, A., Kritis, A., Andreadis, D., Papachristou, E., Leyhausen, G., Koidis, P., Geurtsen, W., & Tsiftsoglou, A. (2015). Angiogenic potential and Secretome of human apical papilla mesenchymal stem cells in various stress microenvironments. Stem Cells and Development, 24(21), 2496–2512. https://doi.org/10.1089/scd.2015.0197
Heo, S. C., Jeon, E. S., Lee, I. H., Kim, H. S., Kim, M. B., & Kim, J. H. (2011). Tumor necrosis factor-α-activated human adipose tissue-derived mesenchymal stem cells accelerate cutaneous wound healing through paracrine mechanisms. Journal of Investigative Dermatology, 131(7), 1559–1567. https://doi.org/10.1038/jid.2011.64
Lu, Z., Wang, G., Dunstan, C. R., Chen, Y., Yenn-Ru Lu, W., Davies, B., & Zreiqat, H. (2013). Activation and promotion of adipose stem cells by tumour necrosis factor-alpha preconditioning for bone regeneration. Journal of Cellular Physiology, 228(8), 1737–1744. https://doi.org/10.1002/jcp.24330
Lee, M. J., Kim, J., Kim, M. Y., Bae, Y. S., Ryu, S. H., Lee, T. G., & Kim, J. H. (2010). Proteomic analysis of tumor necrosis factor-α-induced secretome of human adipose tissue-derived mesenchymal stem cells. Journal of Proteome Research, 9(4), 1754–1762. https://doi.org/10.1021/pr900898n
Miranda, J. P., Camões, S. P., Gaspar, M. M., Rodrigues, J. S., Carvalheiro, M., Bárcia, R. N., Cruz, P., Cruz, H., Simões, S., & Santos, J. M. (2019). The secretome derived from 3D-cultured umbilical cord tissue MSCS counteracts manifestations typifying rheumatoid arthritis. Frontiers in Immunology, 10, 18. https://doi.org/10.3389/fimmu.2019.00018
Kawashima, N. (2012). Characterisation of dental pulp stem cells: A new horizon for tissue regeneration? Archives of Oral Biology, 57(11), 1439–1458. https://doi.org/10.1016/j.archoralbio.2012.08.010
Leyendecker Junior, A., Gomes Pinheiro, C. C., Lazzaretti Fernandes, T., & Franco Bueno, D. (2018). The use of human dental pulp stem cells for in vivo bone tissue engineering: A systematic review. Journal of Tissue Engineering, 9, 204173141775276. https://doi.org/10.1177/2041731417752766
Luo, L., He, Y., Wang, X., Key, B., Lee, B. H., Li, H., & Ye, Q. (2018). Potential roles of dental pulp stem cells in neural regeneration and repair. Stem Cells International. https://doi.org/10.1155/2018/1731289
Yamada, Y., Nakamura-Yamada, S., Kusano, K., & Baba, S. (2019). Clinical potential and current progress of dental pulp stem cells for various systemic diseases in regenerative medicine: A concise review. International Journal of Molecular Sciences, 20(5). https://doi.org/10.3390/ijms20051132
Harrell, C. R., Fellabaum, C., Jovicic, N., Djonov, V., Arsenijevic, N., & Volarevic, V. (2019). Molecular mechanisms responsible for therapeutic potential of mesenchymal stem cell-derived Secretome. Cells, 8(5), 467. https://doi.org/10.3390/cells8050467
de Cara, S. P. H. M., Origassa, C. S. T., de Sá Silva, F., Moreira, M. S. N. A., de Almeida, D. C., Pedroni, A. C. F., Carvalho, G. L., Cury, D. P., Câmara, N. O. S., & Marques, M. M. (2019). Angiogenic properties of dental pulp stem cells conditioned medium on endothelial cells in vitro and in rodent orthotopic dental pulp regeneration. Heliyon, 5(4), e01560. https://doi.org/10.1016/j.heliyon.2019.e01560
Gunawardena, T. N. A., Rahman, M. T., Abdullah, B. J. J., & Abu Kasim, N. H. (2019). Conditioned media derived from mesenchymal stem cell cultures: The next generation for regenerative medicine. Journal of Tissue Engineering and Regenerative Medicine, 13, 569–586. https://doi.org/10.1002/term.2806
Zheng, C., Chen, J., Liu, S., & Jin, Y. (2019). Stem cell-based bone and dental regeneration: A view of microenvironmental modulation. International Journal of Oral Science. Sichuan University Press, 11, 23. https://doi.org/10.1038/s41368-019-0060-3
El Moshy, S., Radwan, I. A., Rady, D., Abbass, M. M. S., El-Rashidy, A. A., Sadek, K. M., … Fawzy El-Sayed, K. M. (2020). Dental stem cell-derivedSecretome/conditioned Medium: The Future for Regenerative Therapeutic Applications. Stem Cells International. https://doi.org/10.1155/2020/7593402
Bakopoulou, A., Papachristou, E., Hadjichristou, C., Bousnaki, M., Theoharidou, A., Kontonasaki, E., … Koidis, P. (2015). Human treated-dentin matrices combined with mg-based scaffolds for dentin regeneration. In IADR General Session BOSTON MASS USA.
Polten, F., Reboll, M. R., Widera, C., Kempf, T., Bethmann, K., Gupta, P., Miglietta, J., Pekcec, A., Tillmanns, J., Bauersachs, J., Giannitsis, E., Pich, A., & Wollert, K. C. (2019). Plasma concentrations of myeloid-derived growth factor in healthy individuals and patients with acute myocardial infarction as assessed by multiple reaction monitoring-mass spectrometry. Analytical Chemistry, 91(2), 1302–1308. https://doi.org/10.1021/acs.analchem.8b03041
Junemann, J., Lämmerhirt, C. M., Polten, F., Just, I., Gerhard, R., Genth, H., & Pich, A. (2017). Quantification of small GTPase glucosylation by clostridial glucosylating toxins using multiplexed MRM analysis. Proteomics, 17(9). https://doi.org/10.1002/pmic.201700016
Picotti, P., & Aebersold, R. (2012). Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nature Methods, 9, 555–566. https://doi.org/10.1038/nmeth.2015
Zeiser, J., Gerhard, R., Just, I., & Pich, A. (2013). Substrate specificity of clostridial glucosylating toxins and their function on colonocytes analyzed by proteomics techniques. Journal of Proteome Research, 12(4), 1604–1618. https://doi.org/10.1021/pr300973q
Cox, J., & Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology, 26(12), 1367–1372. https://doi.org/10.1038/nbt.1511
Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., & Mann, M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular and Cellular Proteomics, 13(9), 2513–2526. https://doi.org/10.1074/mcp.M113.031591
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., Mann, M., & Cox, J. (2016, August 30). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods. Nature Publishing Group, 13, 731–740. https://doi.org/10.1038/nmeth.3901
http://geneontology.org/. (n.d.).
http://www.pantherdb.org/tools/. (n.d.).
Carbon, S., Dietze, H., Lewis, S. E., Mungall, C. J., Munoz-Torres, M. C., Basu, S., et al. (2017). Expansion of the gene ontology knowledgebase and resources: The gene ontology consortium. Nucleic Acids Research, 45(D1), D331–D338. https://doi.org/10.1093/nar/gkw1108
https://reactome.org/. (n.d.).
Croft, D., O’Kelly, G., Wu, G., Haw, R., Gillespie, M., Matthews, L., et al. (2011). Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Research, 39(SUPPL. 1), 691–697. https://doi.org/10.1093/nar/gkq1018
https://string-db.org/. (n.d.).
Caccia, D., Dugo, M., Callari, M., & Bongarzone, I. (2013). Biochimica et Biophysica Acta bioinformatics tools for secretome analysis ☆. BBA - Proteins and Proteomics, 1834(11), 2442–2453. https://doi.org/10.1016/j.bbapap.2013.01.039
Pacienza, N., Lee, R. H., Bae, E. H., Kim, D. K., Liu, Q., Prockop, D. J., & Yannarelli, G. (2019). In vitro macrophage assay predicts the in vivo anti-inflammatory potential of exosomes from human mesenchymal stromal cells. Molecular Therapy - Methods and Clinical Development, 13(June), 67–76. https://doi.org/10.1016/j.omtm.2018.12.003
Vizoso, F. J., Eiro, N., Cid, S., Schneider, J., & Perez-Fernandez, R. (2017). Mesenchymal stem cell secretome: Toward cell-free therapeutic strategies in regenerative medicine. International Journal of Molecular Sciences, 18(9). https://doi.org/10.3390/ijms18091852
Karpievitch, Y. V., Polpitiya, A. D., Anderson, G. A., Smith, R. D., & Dabney, A. R. (2010). Liquid chromatography mass spectrometry-based proteomics: Biological and technological aspects. Annals of Applied Statistics, 4(4), 1797–1823. https://doi.org/10.1214/10-AOAS341
Weitzel, K., Chemie, F., Rev, M. S., Introduction, I., & Reference, C. (2011). Liquid chromatography – tandem mass spectrometry applications in endocrinology. WHO Library Cataloguing-in-Publication Data, (i), 221–235. https://doi.org/10.1002/mas.
Gonzales, P. A., Pisitkun, T., Hoffert, J. D., Tchapyjnikov, D., Star, R. A., Kleta, R., Wang, N. S., & Knepper, M. A. (2009). Large-scale proteomics and phosphoproteomics of urinary exosomes. Journal of the American Society of Nephrology, 20(2), 363–379. https://doi.org/10.1681/ASN.2008040406
Principe, S., Jones, E. E., Kim, Y., Sinha, A., Nyalwidhe, J. O., Brooks, J., Semmes, O. J., Troyer, D. A., Lance, R. S., Kislinger, T., & Drake, R. R. (2013). In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics, 13(10–11), 1667–1671. https://doi.org/10.1002/pmic.201200561
Ettinger, A., & Wittmann, T. (2014). Fluorescence live cell imaging. In: Methods in Cell Biology (Vol. 123). https://doi.org/10.1016/B978-0-12-420138-5.00005-7.
Madrigal, M., Rao, K. S., & Riordan, N. H. (2014). A review of therapeutic effects of mesenchymal stem cell secretions and induction of secretory modification by different culture methods. Journal of Translational Medicine, 12(1), 1–14. https://doi.org/10.1186/s12967-014-0260-8
Bakopoulou, A., & About, I. (2016). Stem cells of dental origin: current research trends and key milestones towards clinical application. Stem Cells International. https://doi.org/10.1155/2016/4209891
Kshitiz, Ellison, D. D., Suhail, Y., Afzal, J., Woo, L., Kilic, O., … Levchenko, A. (2019). Dynamic secretome of bone marrow-derived stromal cells reveals a cardioprotective biochemical cocktail. Proceedings of the National Academy of Sciences of the United States of America, 116(28), 14374–14383. https://doi.org/10.1073/pnas.1902598116.
Ahmed, N. E. M. B., Murakami, M., Kaneko, S., & Nakashima, M. (2016). The effects of hypoxia on the stemness properties of human dental pulp stem cells (DPSCs). Scientific Reports, 6(September), 1–10. https://doi.org/10.1038/srep35476
Dou, L., Yan, Q., Liang, P., Zhou, P., Zhang, Y., & Ji, P. (2018). iTRAQ-based proteomic analysis exploring the influence of hypoxia on the proteome of dental pulp stem cells under 3D culture. Proteomics, 18(3–4), 1–9. https://doi.org/10.1002/pmic.201700215
Hall, B. E., Zhang, L., Sun, Z. J., Utreras, E., Prochazkova, M., Cho, A., Terse, A., Arany, P., Dolan, J. C., Schmidt, B. L., & Kulkarni, A. B. (2016). Conditional TNF-α overexpression in the tooth and alveolar bone results in painful pulpitis and osteitis. Journal of Dental Research, 95(2), 188–195. https://doi.org/10.1177/0022034515612022
Kellesarian, S. V., Al-Kheraif, A. A., Vohra, F., Ghanem, A., Malmstrom, H., Romanos, G. E., & Javed, F. (2016). Cytokine profile in the synovial fluid of patients with temporomandibular joint disorders: A systematic review. Cytokine, 77, 98–106. https://doi.org/10.1016/j.cyto.2015.11.005
Chen, H., Min, X. H., Wang, Q. Y., Leung, F. W., Shi, L., Zhou, Y., Yu, T., Wang, C. M., An, G., Sha, W. H., & Chen, Q. K. (2015). Pre-activation of mesenchymal stem cells with TNF-α, IL-1β 2 and nitric oxide enhances its paracrine effects on radiation-induced intestinal injury. Scientific Reports, 5, 1–14. https://doi.org/10.1038/srep08718
Kwon, Y. W., Heo, S. C., Jeong, G. O., Yoon, J. W., Mo, W. M., Lee, M. J., Jang, I. H., Kwon, S. M., Lee, J. S., & Kim, J. H. (2013). Tumor necrosis factor-α-activated mesenchymal stem cells promote endothelial progenitor cell homing and angiogenesis. Biochimica et Biophysica Acta - Molecular Basis of Disease, 1832(12), 2136–2144. https://doi.org/10.1016/j.bbadis.2013.08.002
Wajant, H., Pfizenmaier, K., & Scheurich, P. (2003). Tumor necrosis factor signaling. Cell Death and Differentiation, 10(1), 45–65. https://doi.org/10.1038/sj.cdd.4401189
Wojdasiewicz, P., Poniatowski, Ł. A., & Szukiewicz, D. (2014). The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediators of Inflammation, 2014, 1–19. https://doi.org/10.1155/2014/561459
Ighodaro, O. M., & Akinloye, O. A. (2018). First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alexandria Journal of Medicine, 54(4), 287–293. https://doi.org/10.1016/j.ajme.2017.09.001
Matsumoto, H., Silverton, S. F., Debolt, K., & Shapiro, I. M. (1991). Superoxide dismutase and catalase activities in the growth cartilage: Relationship between oxidoreductase activity and chondrocyte maturation. Journal of Bone and Mineral Research, 6(6), 569–574. https://doi.org/10.1002/jbmr.5650060607
Maines, M. D. (1997). The heme oxygenase system: A regulator of second messenger gases. Annual Review of Pharmacology and Toxicology, 37(1), 517–554. https://doi.org/10.1146/annurev.pharmtox.37.1.517
Khan, N. M., Ahmad, I., & Haqqi, T. M. (2018). Nrf2/ARE pathway attenuates oxidative and apoptotic response in human osteoarthritis chondrocytes by activating ERK1/2/ELK1-P70S6K-P90RSK signaling axis. Free Radical Biology and Medicine, 116(August 2017), 159–171. https://doi.org/10.1016/j.freeradbiomed.2018.01.013
Ahmed, S. M. U., Luo, L., Namani, A., Wang, X. J., & Tang, X. (2017). Nrf2 signaling pathway: Pivotal roles in inflammation. Biochimica et Biophysica Acta - Molecular Basis of Disease. Elsevier B.V. https://doi.org/10.1016/j.bbadis.2016.11.005
Kong, P., Chen, G., Jiang, A., Wang, Y., Song, C., Zhuang, J., … Yan, J. (2016). Sesamin inhibits IL-1β-stimulated inflammatory response in human osteoarthritis chondrocytes by activating Nrf2 signaling pathway. Oncotarget, 7(50), 83720–83726. https://doi.org/10.18632/oncotarget.13360.
Khan, N. M., Haseeb, A., Ansari, M. Y., Devarapalli, P., Haynie, S., & Haqqi, T. M. (2017). Wogonin, a plant derived small molecule, exerts potent anti-inflammatory and chondroprotective effects through the activation of ROS/ERK/Nrf2 signaling pathways in human osteoarthritis chondrocytes. Free Radical Biology and Medicine, 106, 288–301. https://doi.org/10.1016/j.freeradbiomed.2017.02.041
Xu, X., Zheng, L., Yuan, Q., Zhen, G., Crane, J. L., Zhou, X., & Cao, X. (2018). Transforming growth factor-β in stem cells and tissue homeostasis. Bone Research, 6(1), 2. https://doi.org/10.1038/s41413-017-0005-4
Tang, X., Fan, L., Pei, M., Zeng, L., & Ge, Z. (2015). Evolving concepts of chondrogenic differentiation: History, state-of-the-art and future perspectives. European Cells & Materials, 30, 12–27. https://doi.org/10.22203/eCM.v030a02
Brew, K., & Nagase, H. (2010). The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. Biochimica et Biophysica Acta, Molecular Cell Research, 1803(1), 55–71. https://doi.org/10.1016/j.bbamcr.2010.01.003
Dean, D. D., Martel-Pelletier, J., Pelletier, J. P., Howell, D. S., & Woessner, J. F. (1989). Evidence for metalloproteinase inhibitor imbalance in human osteoarthritic cartilage. Journal of Clinical Investigation, 84(2), 678–685. https://doi.org/10.1172/JCI114215
Pelletier, J. -P., Mineau, F., Faure, M. -P., & Martel-Pelletier, J. (1990). Imbalance between the mechanisms of activation and inhibition of metalloproteases in the early lesions of experimental osteoarthritis. Arthritis and Rheumatism, 33(10), 1466–1476. https://doi.org/10.1002/art.1780331003
Nakashima, M., Iohara, K., & Sugiyama, M. (2009). Human dental pulp stem cells with highly angiogenic and neurogenic potential for possible use in pulp regeneration. Cytokine and Growth Factor Reviews, 20(5–6), 435–440. https://doi.org/10.1016/j.cytogfr.2009.10.012
Ishizaka, R., Hayashi, Y., Iohara, K., Sugiyama, M., Murakami, M., Yamamoto, T., Fukuta, O., & Nakashima, M. (2013). Stimulation of angiogenesis, neurogenesis and regeneration by side population cells from dental pulp. Biomaterials, 34(8), 1888–1897. https://doi.org/10.1016/j.biomaterials.2012.10.045
Bronckaers, A., Hilkens, P., Fanton, Y., Struys, T., Gervois, P., Politis, C., Martens, W., & Lambrichts, I. (2013). Angiogenic properties of human dental pulp stem cells. PLoS One, 8(8). https://doi.org/10.1371/journal.pone.0071104
Hilkens, P., Bronckaers, A., Ratajczak, J., Gervois, P., Wolfs, E., & Lambrichts, I. (2017). The Angiogenic potential of DPSCs and SCAPs in an in vivo model of dental pulp regeneration. Stem Cells International, 2017, 18–22. https://doi.org/10.1155/2017/2582080
Zhou, Y., Yamamoto, Y., Xiao, Z., & Ochiya, T. (2019). The immunomodulatory functions of mesenchymal stromal/stem cells mediated via paracrine activity. Journal of Clinical Medicine, 8(7), 1025. https://doi.org/10.3390/jcm8071025
Davies, L. C., Jenkins, S. J., Allen, J. E., & Taylor, P. R. (2013). Tissue-resident macrophages. Nature Immunology, 14(10), 986–995. https://doi.org/10.1038/ni.2705
Kellum, J. A., Song, M., & Li, J. (2004). Lactic and hydrochloric acids induce different patterns of inflammatory response in LPS-stimulated RAW 264.7 cells. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 286(4), 686–692. https://doi.org/10.1152/ajpregu.00564.2003
Soromou, L. W., Zhang, Z., Li, R., Chen, N., Guo, W., Huo, M., Guan, S., Lu, J., & Deng, X. (2012). Regulation of inflammatory cytokines in lipopolysaccharide-stimulated RAW 264.7 murine macrophage by 7-O-methyl-naringenin. Molecules, 17(3), 3574–3585. https://doi.org/10.3390/molecules17033574
Murphy, G., & Nagase, H. (2008). Reappraising metalloproteinases in rheumatoid arthritis and osteoarthritis: Destruction or repair? Nature Clinical Practice Rheumatology, 4(3), 128–135. https://doi.org/10.1038/ncprheum0727
Yamamoto, K., Murphy, G., & Troeberg, L. (2015). Extracellular regulation of metalloproteinases. Matrix Biology, 44–46, 255–263. https://doi.org/10.1016/j.matbio.2015.02.007
Ishimaru, J. I., Oguma, Y., & Goss, A. N. (2000). Matrix metalloproteinase and tissue inhibitor of metalloproteinase in serum and lavage synovial fluid of patients with temporomandibular joint disorders. British Journal of Oral and Maxillofacial Surgery, 38(4), 354–359. https://doi.org/10.1054/bjom.2000.0306
Tiilikainen, P., Pirttiniemi, P., Kainulainen, T., Pernu, H., & Raustia, A. (2005). MMP-3 and -8 expression is found in the condylar surface of temporomandibular joints with internal derangement. Journal of Oral Pathology and Medicine, 34(1), 39–45. https://doi.org/10.1111/j.1600-0714.2004.00262.x
Almeida, L. E., Caporal, K., Ambros, V., Azevedo, M., Noronha, L., Leonardi, R., & Trevilatto, P. C. (2015). Immunohistochemical expression of matrix metalloprotease-2 and matrix metalloprotease-9 in the disks of patients with temporomandibular joint dysfunction. Journal of Oral Pathology and Medicine, 44(1), 75–79. https://doi.org/10.1111/jop.12213
van Buul, G. M., Villafuertes, E., Bos, P. K., Waarsing, J. H., Kops, N., Narcisi, R., Weinans, H., Verhaar, J. A. N., Bernsen, M. R., & van Osch, G. J. V. M. (2012). Mesenchymal stem cells secrete factors that inhibit inflammatory processes in short-term osteoarthritic synovium and cartilage explant culture. Osteoarthritis and Cartilage, 20(10), 1186–1196. https://doi.org/10.1016/j.joca.2012.06.003
Platas, J., Guillén, M. I., Del Caz, M. D. P., Gomar, F., Mirabet, V., & Alcaraz, M. J. (2013). Conditioned media from adipose-tissue-derived mesenchymal stem cells downregulate degradative mediators induced by interleukin-1 β in osteoarthritic chondrocytes. Mediators of Inflammation. https://doi.org/10.1155/2013/357014
Iyer, S. S., & Cheng, G. (2012). Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Critical Reviews in Immunology, 32(1), 23–63. https://doi.org/10.1615/critrevimmunol.v32.i1.30
Ti, D., Hao, H., Tong, C., Liu, J., Dong, L., Zheng, J., Zhao, Y., Liu, H., Fu, X., & Han, W. (2015). LPS-preconditioned mesenchymal stromal cells modify macrophage polarization for resolution of chronic inflammation via exosome-shuttled let-7b. Journal of Translational Medicine, 13(1), 308. https://doi.org/10.1186/s12967-015-0642-6
Acknowledgments
We would like to thank Karsten Heidrich and Ulrike Schrameck, from the Research Core Unit Proteomics & Institute of Toxicology, Hannover Medical School, for good technical assistance.
Code Availability
Not applicable to this study.
Funding
This research is co-financed by Greece and the European Union (European Social Fund- ESF) through the Operational Programme «Human Resources Development, Education and Lifelong Learning» in the context of the project “Strengthening Human Resources Research Potential via Doctorate Research” (MIS-5000432), implemented by the State Scholarships Foundation (ΙΚΥ).
Author information
Authors and Affiliations
Contributions
MB: Methodology, Validation, Formal analysis, Investigation, Writing - Original Draft, Visualization. AB: Conceptualization, Resources, Writing – Review & Editing, Supervision, Project administration, Funding acquisition. AP: Methodology, Software, Formal analysis, Writing – Review & Editing. EP: Methodology, Resources. AK: Resources, Writing – Review & Editing. PK: Conceptualization, Writing – Review & Editing, Supervision, Project administration, Funding acquisition.
Corresponding authors
Ethics declarations
Ethics Approval
The study was approved by the Institutional Ethical Review Board (Protocol No. 20/29–11–2017).
Consent to Participate
All patients signed an informed consent form.
Consent for Publication
All authors have given their consent for publication and have reviewed and approved the submission.
Conflict of Interest
Authors declare that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Fig. 1
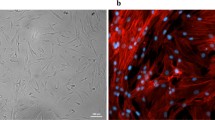
Characterization of dental pulp stem cells (DPSCs). Characterization of DPSCs by flow cytometry for the expression of mesenchymal (CD90/Thy-1, CD73, CD49f/a6-integrin, CD146/MUC18, STRO-1, CD34), endothelial (CD105/endoglin,) hematopoietic (CD45) and embryonic (SSEA-4) stem (SC) markers (green line: unstained control, red line: marker of interest). Results are means of triplicates (± SD) of two independent experiments. (PNG 5454 kb)
Supplementary Fig. 2
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) of conditioned medium (CM) (a & b) and cell lysate (CL) (c) samples. a & b. Proteins of CM samples were separated by SDS-PAGE on a 10% acrylamide gel and then stained with Coomassie Brilliant blue. Lane 1 depicts the protein markers, while lanes 2 to 5 depict the CM samples as follows: CM_Norm, CM_Norm+TNF-α, CM_Hyp, CM_Hyp + TNF-α. c. Proteins of CL samples were separated by SDS-PAGE electrophoresis on a 15% acrylamide gel and then stained with Coomassie Brilliant blue. Lane 1 depicts the protein markers, while lanes 2 to 5 depict the CL samples as follows: CL_Norm, CL_Norm+TNF-α, CL_Hyp, CL_Hyp + TNF-α. (PNG 4718 kb)
Supplementary Fig. 3
Gene Ontology (GO) analysis of the CM samples regarding the term “Cellular Compartment”. (PNG 14148 kb)
Supplementary Fig. 4
Gene Ontology (GO) analysis of the CM-CL samples comparisons regarding the term “Cellular Compartment” (a-d), “Biological Process” (e-h), and “Pathway” (i-l). (PNG 12659 kb)
ESM 1
(PNG 12832 kb)
ESM 2
(PNG 12988 kb)
Supplementary Fig. 5
Reactome Pathway analysis of CM-CL samples comparisons for a wide range of pathways (a-d) and specific pathways regarding the immune system (e-h). (PNG 12629 kb)
ESM 3
(PNG 12752 kb)
Supplementary Fig. 6A
Viability/proliferation of RAW cells after the application of LPS and CM. The immune cell response induced by CM was tested initially assessed with an MTT assay, which showed a significant increase in cell viability/proliferation 24 h after LPS application. Addition of CM in LPS-stimulated RAW cells resulted in a decrease viability/proliferation. This effect was reversed after 48 h, where addition of CM samples had a stimulatory effect, resulting in significant increase in cell viability compared to control and LPS-stimulated RAW cells. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. 6B Morphological assessment of RAW cells after the application of LPS and CM samples. Normal RAW cells (a), RAW cells+LPS (b), RAW cells+LPS + DMEM 24 h (c) and 48 h (d), RAW cells+LPS + CM_Norm 24 h (e), RAW cells+LPS + CM_Norm+TNF-α 24 h (f), RAW cells+LPS + CM_Hyp 24 h (g), RAW cells+LPS + CM_Hyp + TNF-α 24 h (h), RAW cells+LPS + CM_Norm 48 h (i), RAW cells+LPS + CM_Norm+TNF-α 48 h (j), RAW cells+LPS + CM_Hyp 48 h (k), and RAW cells+LPS + CM_Hyp + TNF-α 48 h (l). Normal RAW cells display an irregular form with pseudopodia (a). LPS application affected the morphology of RAW cells (b), which were transformed into a totally circular form after 24 and 48 h of the LPS application (c and d). This effect was partly reversed by the application of CM samples after 24 h (e-h), while the effect of CM_Norm was evident even 48 h after culture (i). (PNG 6137 kb)
ESM 4
(PNG 14033 kb)
Supplementary Table 1
(DOCX 183 kb)
Rights and permissions
About this article
Cite this article
Bousnaki, ., Bakopoulou, A., Pich, A. et al. Mapping the Secretome of Dental Pulp Stem Cells Under Variable Microenvironmental Conditions. Stem Cell Rev and Rep 18, 1372–1407 (2022). https://doi.org/10.1007/s12015-021-10255-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-021-10255-2