Abstract
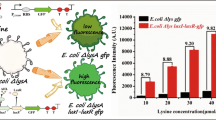
Detection of 2,4,6-trinitrotoluene (TNT) has been extensively studied since it is a common explosive filling for landmines, posing significant threats to the environment and human safety. The rapid advances in synthetic biology give new hope to detect such toxic and hazardous compounds in a more sensitive and safe way. Biosensor construction anticipates finding sensing elements able to detect TNT. As TNT can induce some physiological responses in E. coli, it may be useful to define the sensing elements from E. coli to detect TNT. An E. coli MG1655 genomic promoter library containing nearly 5,400 elements was constructed. Five elements, yadG, yqgC, aspC, recE, and topA, displayed high sensing specificity to TNT and its indicator compounds 1,3-DNB and 2,4-DNT. Based on this, a whole cell biosensor was constructed using E. coli, in which green fluorescent protein was positioned downstream of the five sensing elements via genetic fusion. The threshold value, detection time, EC200 value, and other aspects of five sensing elements were determined and the minimum responding concentration to TNT was 4.75 mg/L. According to the synthetic biology, the five sensing elements enriched the reservoir of TNT-sensing elements, and provided a more applicable toolkit to be applied in genetic routes and live systems of biosensors in future.






Similar content being viewed by others
References
Berthe-Corti, L., Jacobi, H., Kleihauer, S., & Witte, I. (1998). Cytotoxicity and mutagenicity of a 2,4,6-trinitrotoluene (TNT) and hexogen contaminated soil in S. typhimurium and mammalian cells. Chemosphere, 37, 209–218.
Reddy, G., Reddy, T. V., Choudhury, H., Daniel, F. B., & Leach, G. J. (1997). Assessment of environmental hazards of 1,3,5-trinitrobenzene. Journal of Toxicology and Environment Health, 52, 447–460.
U.S. Environmental Protection Agency. (1996). Innovative treatment technologies: Annual status report (8th ed.). Washington, DC: U.S. Environmental Protection Agency. Retrieved Novermber 8, 1996 from http://nepis.epa.gov/Exe/ZyNET.exe/10002Y79.TXT?ZyActionD=ZyDocument&Client=EPA&Index=1995+Thru+1999&Docs=&Query=&Time=&EndTime=&SearchMethod=1&TocRestrict=n&Toc=&TocEntry=&QField=&QFieldYear=&QFieldMonth=&QFieldDay=&IntQFieldOp=0&ExtQFieldOp=0&XmlQuery=&File=D%3A%5Czyfiles%5CIndex%20Data%5C95thru99%5CTxt%5C00000004%5C10002Y79.txt&User=ANONYMOUS&Password=anonymous&SortMethod=h%7C-&MaximumDocuments=1&FuzzyDegree=0&ImageQuality=r75g8/r75g8/x150y150g16/i425&Display=p%7Cf&DefSeekPage=x&SearchBack=ZyActionL&Back=ZyActionS&BackDesc=Results%20page&MaximumPages=1&ZyEntry=1&SeekPage=x&ZyPURL.
Frische, T. (2002). Screening for soil toxicity and mutagenicity using luminescent bacteria—A case study of the explosive 2,4,6-trinitrotoluene (TNT). Ecotoxicology and Environmental Safety, 51(2), 133–144.
Resolution adopted by the General Assembly on 8 December 2005 on the report of the Special Political and Decolonization Committee (Fourth Committee) (A/60/473), sixtieth session Agenda item 27, Distr.: General 18 January 2006.
Benner, S. A., & Sismour, A. M. (2005). Synthetic biology. Nature Reviews Genetics, 6(7), 533–543.
Leedjarv, A., Ivask, A., Virta, M., & Kahru, A. (2006). Analysis of bioavailable phenols from natural samples by recombinant luminescent bacterial sensors. Chemosphere, 11, 1910–1919.
Magrisso, S., Erel, Y., & Belkin, S. (2008). Microbial reporters of metal bioavailability. Microbial Biotechnology, 1, 320–330.
Robin, T., & van der Meer, J. R. (2008). Bacterial biosensors for measuring availability of environmental pollutants. Sensors, 8, 4062–4080.
Trang, P. T. K., Berg, M., Viet, P. H., Mui, N. V., & van der Meer, J. R. (2005). Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environmental Science and Technology, 19, 7625–7630.
Werlen, C., Jaspers, M. C. M., & van der Meer, J. R. (2004). Measurement of biologically available naphthalene in gas, and aqueous phases by use of a Pseudomonas putida biosensor. Applied and Environment Microbiology, 70, 43–51.
Meighen, E. A., & Szittner, R. B. (1992). Multiple repetitive elements andorganization of the lux operons of luminescent terrestrial bacteria. Journal of Bacteriology, 174, 5371–5381.
Kurvet, I., Ivask, A., Bondarenko, O., Sihtmäe, M., & Kahru, A. (2011). LuxCDABE-transformed constitutively bioluminescent Escherichia coli for toxicity screening: Comparison with naturally luminous Vibrio fischeri. Sensors, 11(8), 7865–7878.
van der Meer, J. R., & Belkin, S. (2010). Where microbiology meets microengineering: Design and applications of reporter bacteria. Nature Reviews Microbiology, 8(7), 511–522.
Burlage, R., Youngblood, T., & Lamothe, D. (1998). Bioreporter bacteria for landmine detection. RNL report/CP-96972. Oak Ridge: Oak Ridge National Laboratory.
Burlage, R., Patek, D., & Everman, K. (1999). Method for detection of buried explosives using a biosensor. US Patent 5:972,638.
Kyoko, T., Sumio, G., Kiwao, K., Masana, H., Takao, I., & Manabu, S. (2001). Modification of umu Test using the bioluminescent bacteria and application to sediments and soils. Journals in Environmental Chemistry, 11(4), 841–848.
Garmendia, J., De Las Heras, A., Galvão, T., & De Lorenzo, V. (2008). Tracing explosives in soil with transcriptional regulators of Pseudomonas putida evolved for responding to nitrotoluenes. Microbial Biotechnology, 1(3), 236–246.
González-Pérez, M., Van Dillewijn, P., Wittich, R., & Ramos, J. (2007). Escherichia coli has multiple enzymes that attack TNT and release nitrogen for growth. Environmental Microbiology, 9(6), 1535–1540.
Behzadian, F., Barjeste, H., Hosseinkhani, S., & Zarei, A. R. (2012). Zarei construction and characterization of Escherichia coli whole-cell biosensors for toluene and related compounds. Current Microbiology, 62, 690–696.
Sharon, Y., Chaim, L., Rachel, R., Neta, B., Yaara, M., & Shimshon, B. (2014). Escherichia coli bioreporters for the detection of 2,4-dinitrotoluene and 2,4,6-trinitrotoluene. Applied Microbiology and Biotechnology, 98(2), 885–895.
Miller, W., Leveau, J., & Lindow, S. (2000). Improved Gfp and inaZ broad-host-range promoter-probe vectors. Molecular Plant-Microbe Interactions, 13, 1243–1250.
Lerner, C. G., & Inouye, M. (1990). Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Research, 18, 4631.
Wang, R. F., & Kushner, S. R. (1991). Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene, 100, 195–199.
Belkin, S., Smulski, D., Dadon, S., Vollmer, A., Van Dyk, T., & Larossa, R. (1997). A panel of stress-responsive luminous bacteria for the detection of selected classes of toxicants. Water Research, 31(12), 3009–3016.
Belkin, S. (1998). A panel of stress-responsive luminous bacteria for monitoring wastewater toxicity. Methods in Molecular Biology, 102, 247–258.
Nishino, S. F., Spain, J. C., Lenke, H., et al. (1999). Mineralizat ion of 2,4- and 2,6-DNT in soil slurries. Environmental Science and Technology, 33, 1060–1064.
Habib, M. (2007). Controlled biological and biomimetic systems for landmine detection. Biosensors & Bioelectronics, 23(1), 1–18.
Gillen, J. R., Willis, D. K., & Clark, A. J. (1981). Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. Journal of Bacteriology, 145, 521–532.
Yang, Y., & Fix, D. (2006). Genetic analysis of the anti-mutagenic effect of genistein in Escherichia coli. Mutation Research, 600(1–2), 193–206.
Tse-Dinh, Y.-C., & Wang, J. C. (1986). Complete nucleotide sequence of the topA gene encoding Escherichia coli DNA topoisomerase I. Journal of Molecular Biology, 191(3), 321–331.
Rao, M. R., & Halfhill, M. D. (2009). Phytoremediation and phytosensing of chemical contaminants, RDX and TNT: identification of the required target genes. Functional & Integrative Genomics, 9(4), 537–547.
Pomeranz, M. C. (2005). ABC promoter up regulation using TNT and Kangenyacin. University of Tennessee Honors Thesis Projects. RDX and TNT: Identification of the required target genes. Functional & Integrative Genomics, 9(4), 537–547.
Seiki, K., Sachiko, O., Tomoko, O., Hideyuki, O., & Hiroyuki, K. (1990). Aspartate aminotransferase of Escherichia coli: Nucleotide sequence of the aspc gene. Journal of Biochemistry, 97(4), 1259–1262.
Martin, W., Ulrich, T., Nicole, R., Christoph, K., & Stefan, S. (2012). Future security. Communications in Computer and Information Science, 318, 432–437.
De Las, Heras. A., Carreño, C., & De Lorenzo, V. (2008). Stable implantation of orthogonal sensor circuits in gram-negative bacteria for environmental release. Environmental Microbiology, 10(12), 3305–3316.
Gibbs, W. W. (2004). Synthetic life. Scientific American, 290(5), 48–55.
Endy, D. (2005). Foundations for engineering biology. Nature, 438, 449–453.
Blattner, F., Plunkett, G., Bloch, C., Perna, N., Burland, V., Riley, M., et al. (1997). Thecomplete genome sequence of Escherichia coli K-12. Science, 277(5331), 1453–1462.
Grant, S., Jessee, J., Bloom, F., & Hanahan, D. (1990). Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylationrestriction mutants. Proceedings of the National Academy of Sciences of the United States of America, 87(12), 4645–4649.
Acknowledgments
We would like to thank Professor Wang Hengliang (Beijing Institute of Bioengineering) for help and guidance in this study. This study was funded by the synthetic biology project of Chinese National High Technology Research and Development Program (863 Program). Project Number 2012AA02A706.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Tan, J., Kan, N., Wang, W. et al. Construction of 2,4,6-Trinitrotoluene Biosensors with Novel Sensing Elements from Escherichia coli K-12 MG1655. Cell Biochem Biophys 72, 417–428 (2015). https://doi.org/10.1007/s12013-014-0481-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-014-0481-8