Abstract
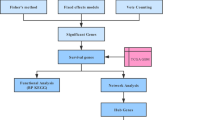
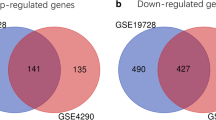
Glioblastoma (GBM) is the most common primary intracranial malignancy with a very low survival rate. Exploring key molecular markers for GBM can help with early diagnosis, prognostic prediction, and recurrence monitoring. This study aims to explore novel biomarkers for GBM via bioinformatics analysis and experimental verification. Dataset GSE103229 was obtained from the GEO database to search differentially expressed lncRNA (DELs), mRNAs (DEMs), and miRNAs (DEMis). Hub genes were selected to establish competing endogenous RNA (ceRNA) networks. The GEPIA database was employed for the survival analysis and expression detection of hub genes. Hub gene expression in GBM tissue samples and cell lines was validated using RT-qPCR. Western blotting was employed for protein expression evaluation. SYT1 overexpression vector was transfected in GBM cells. CCK-8 assay and flow cytometry were performed to detect the malignant phenotypes of GBM cells. There were 901 upregulated and 1086 downregulated DEMs identified, which were prominently enriched in various malignancy-related functions and pathways. Twenty-two hub genes were selected from PPI networks. Survival analysis and experimental validation revealed that four hub genes were tightly associated with GBM prognosis and progression, including SYT1, GRIN2A, KCNA1, and SYNPR. The four genes were significantly downregulated in GBM tissues and cell lines. Overexpressing SYT1 alleviated the proliferation and promoted the apoptosis of GBM cells in vitro. We identify four genes that may be potential molecular markers of GBM, which may provide new ideas for improving early diagnosis and prediction of the disease.











Similar content being viewed by others
Data Availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Shi, Z., et al. (2023). Comprehensive analysis of oxidative stress-related lncRNA signatures in glioma reveals the discrepancy of prognostic and immune infiltration. Science and Reports, 13(1), 7731.
Pirlog, B. O., et al. (2023). New perspective on DNA response pathway (DDR) in glioblastoma, focus on classic biomarkers and emerging roles of ncRNAs. Expert Reviews in Molecular Medicine, 25, e18.
Jafari, M., & Hasanzadeh, M. (2020). Cell-specific frequency as a new hallmark to early detection of cancer and efficient therapy: Recording of cancer voice as a new horizon. Biomedicine & Pharmacotherapy, 122, 109770.
Huang, X., et al. (2018). High throughput single cell RNA sequencing, bioinformatics analysis and applications. Advances in Experimental Medicine and Biology, 1068, 33–43.
Yadav, D.K., et al. (2023). Identification of hub genes associated with prognosis of lung cancer via integrated bioinformatics and in vitro approach. Journal of Biomology Structure and Dynamics, 41(20), 11204–11218.
Yin, X., et al. (2022). Identification of novel prognostic targets in glioblastoma using bioinformatics analysis. Biomedical Engineering Online, 21(1), 26.
Kong, Y., et al. (2020). Identification of immune-related genes contributing to the development of glioblastoma using weighted gene co-expression network analysis. Frontiers in Immunology, 11, 1281.
Geng, R. X., et al. (2018). Identification of core biomarkers associated with outcome in glioma: evidence from bioinformatics analysis. Disease Markers, 2018, 3215958.
Qi, X., et al. (2015). ceRNA in cancer: Possible functions and clinical implications. Journal of Medical Genetics, 52(10), 710–718.
Xiao, H., Liang, S., & Wang, L. (2020). Competing endogenous RNA regulation in hematologic malignancies. Clinica Chimica Acta, 509, 108–116.
Rahnama, S., et al. (2021). Identification of dysregulated competing endogenous RNA networks in glioblastoma: A way toward improved therapeutic opportunities. Life Sciences, 277, 119488.
Peng, Q., et al. (2020). Prediction of a competing endogenous RNA co-expression network as a prognostic marker in glioblastoma. Journal of Cellular and Molecular Medicine, 24(22), 13346–13355.
Barrett, T., et al. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids, 41(Database issue), D991-5.
Sherman, B. T., et al. (2022). DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Research, 50, W216–W221.
da Huang, W., Sherman, B. T., & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57.
Szklarczyk, D., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research, 47(D1), D607-d613.
Li, J. H., et al. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res, 42(Database issue), D92-7.
Tang, Z., et al. (2017). GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research, 45(W1), W98-w102.
Feng, S.W., et al. (2023). Exploring the functional roles of telomere maintenance 2 in the tumorigenesis of glioblastoma multiforme and drug responsiveness to temozolomide. International Journal of Molecular Sciences, 24(11), 9256.
Quddusi, D.M. & Bajcinca, N. (2023). Identification of genomic biomarkers and their pathway crosstalks for deciphering mechanistic links in glioblastoma. IET Systems Biology, 17(4), 143–161.
Zhuang, R., et al. (2023). Rab26 restricts insulin secretion via sequestering Synaptotagmin-1. PLoS Biology, 21(6), e3002142.
Riggs, E., et al. (2022). SYT1-associated neurodevelopmental disorder: A narrative review. Children (Basel), 9(10), 1439.
Lu, H., et al. (2019). miRNA-34a suppresses colon carcinoma proliferation and induces cell apoptosis by targeting SYT1. International Journal of Clinical and Experimental Pathology, 12(8), 2887–2897.
Zhang, J., et al. (2024). miRNA-363–3p Hinders proliferation, migration, invasion and autophagy of thyroid cancer cells by controlling SYT1 transcription to affect NF-κB. Endocrine Metabolic & Immune Disorders Drug Targets, 24(1), 153–162
Yang, J., & Yang, Q. (2020). Identification of core genes and screening of potential targets in glioblastoma multiforme by integrated bioinformatic analysis. Frontiers in Oncology, 10, 615976.
Zhou, Y., et al. (2019). Identification of potential biomarkers in glioblastoma through bioinformatic analysis and evaluating their prognostic value. BioMed Research International, 2019, 6581576.
Myers, S.J., et al. (2019). Distinct roles of GRIN2A and GRIN2B variants in neurological conditions. F1000Res, 8, F1000.
D’Mello, S. A., et al. (2014). Evidence that GRIN2A mutations in melanoma correlate with decreased survival. Frontiers Oncology, 3, 333.
Prickett, T. D., et al. (2014). Somatic mutation of GRIN2A in malignant melanoma results in loss of tumor suppressor activity via aberrant NMDAR complex formation. The Journal of Investigative Dermatology, 134(9), 2390–2398.
D'Adamo, M.C., et al. (2020). Kv1.1 Channelopathies: Pathophysiological mechanisms and therapeutic approaches. International Journal of Molecular Science, 21(8), 2935.
Liu, L., et al. (2019). Silencing of KCNA1 suppresses the cervical cancer development via mitochondria damage. Channels (Austin), 13(1), 321–330.
Uhan, S., et al. (2020). Hypermethylated promoters of genes UNC5D and KCNA1 as potential novel diagnostic biomarkers in colorectal cancer. Epigenomics, 12(19), 1677–1688.
Sun, T., et al. (2006). Differential expression of synaptoporin and synaptophysin in primary sensory neurons and up-regulation of synaptoporin after peripheral nerve injury. Neuroscience, 141(3), 1233–1245.
Su, K., et al. (2021). The role of a ceRNA regulatory network based on lncRNA MALAT1 site in cancer progression. Biomedicine & Pharmacotherapy, 137, 111389.
Chen, X., et al. (2015). miR-372 regulates glioma cell proliferation and invasion by directly targeting PHLPP2. Journal of Cellular Biochemistry, 116(2), 225–232.
Funding
None.
Author information
Authors and Affiliations
Contributions
ZCH and ZJC were main designers of this study, ZCH, ZJC, EPS and PY performed the experiments and analyzed the data, ZCH, ZJC, WWC and HQL drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
The study was approved by the Ethics Committee of Lingnan Hospital, branch of The Third Affiliated Hospital of Sun Yat-sen University. Informed consents were obtained from the participants.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, Z., Chen, Z., Song, E. et al. Bioinformatics Analysis and Experimental Validation for Exploring Key Molecular Markers for Glioblastoma. Appl Biochem Biotechnol (2024). https://doi.org/10.1007/s12010-024-04894-7
Accepted:
Published:
DOI: https://doi.org/10.1007/s12010-024-04894-7