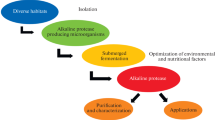
Abstract
The group of hydrolytic enzymes synonymously known as proteases is predominantly most favored for the class of industrial enzymes. The present work focuses on the thermostable nature of these proteolytic enzymes that occur naturally among mesophilic and thermophilic microbes. The broad thermo-active feature (40–80 °C), ease of cultivation, maintenance, and bulk production are the key features associated with these enzymes. Detailing of contemporary production technologies, and controllable operational parameters including the purification strategies, are the key features that justify their industrial dominance as biocatalysts. In addition, the rigorous research inputs by protein engineering and enzyme immobilization studies add up to the thermo-catalytic features and application capabilities of these enzymes. The work summarizes key features of microbial proteases that make them numero-uno for laundry, biomaterials, waste management, food and feed, tannery, and medical as well as pharmaceutical industries. The quest for novel and/or designed and engineered thermostable protease from unexplored sources is highly stimulating and will address the ever-increasing industrial demands.







Similar content being viewed by others
References
Rampelotto, P. H. (2013). Extremophiles and extreme environments. Life., 3(3), 482–485.
Stetter, K. O. (2013). A brief history of the discovery of hyperthermophilic life. Biochemical Society Transactions, 41(1), 416–420.
Raval, V. H., Bhatt, H. B., & Singh, S. P. (2018). Extremophiles (Durvasula, R. V., & Rao, D. S.). CRC Press. US. pp. 137–164
Roca, M., Liu, H., Messer, B., & Warshel, A. (2007). On the relationship between thermal stability and catalytic power of enzymes. Biochem., 46(51), 15076–15088.
Gimenes, N. C., Silveira, E., & Tambourgi, E. B. (2021). An overview of proteases: Production, downstream processes and industrial applications. Separation and Purification Reviews, 50(3), 223–243.
Litchfield, C. D. (2011). Potential for industrial products from the halophilic Archaea. Journal of Industrial Microbiology and Biotechnology, 38(10), 1635–1647.
Klingeberg, M., Hashwa, F., & Antranikian, G. (1991). Properties of extremely thermostable proteases from anaerobic hyperthermophilic bacteria. Applied Microbiology and Biotechnology, 34(6), 715–719.
Toplak, A., Wu, B., Fusetti, F., Quaedflieg, P. J., & Janssen, D. B. (2013). Proteolysin, a novel highly thermostable and cosolvent-compatible protease from the thermophilic bacterium Coprothermobacter proteolyticus. Applied and Environment Microbiology, 79(18), 5625–5632.
Abdollahi, P., Ghane, M., & Babaeekhou, L. (2021). Isolation and characterization of Thermophilic bacteria from Gavmesh Goli hot spring in Sabalan geothermal field, Iran: Thermomonas hydrothermalis and Bacillus altitudinis isolates as a potential source of Thermostable Protease. Geomicrobiology Journal, 38(1), 87–95.
Abu-Khudir, R., Salem, M. M., Allam, N. G., & Ali, E. (2019). Production, Partial purification, and biochemical characterization of a thermotolerant alkaline metallo-protease from Staphylococcus sciuri. Applied Biochemistry and Biotechnology, 189(1), 87–102.
Raval, V. H., Pillai, S., Rawal, C. M., & Singh, S. P. (2014). Biochemical and structural characterization of a detergent-stable serine alkaline protease from seawater haloalkaliphilic bacteria. Process Biochemistry, 49(6), 955–962.
Al-Dhabi, N. A., Esmail, G. A., Ghilan, A. K. M., Arasu, M. V., Duraipandiyan, V., & Ponmurugan, K. (2020). Characterization and fermentation optimization of novel thermo stable alkaline protease from Streptomyces sp. Al-Dhabi-82 from the Saudi Arabian environment for eco-friendly and industrial applications. J. King Saud Univ. Sci., 32(1), 1258–1264.
Gemechu, G., Masi, C., Tafesse, M., & Kebede, G. (2020). A review on the bacterial alkaline proteases. J Xidian Univ., 14(11), 632–634.
Katchalski-Katzir, E., & Kraemer, D. M. (2000). Eupergit® C, a carrier for immobilization of enzymes of industrial potential. Journal of Molecular Catalysis. B, Enzymatic, 10(1–3), 157–176.
Singh, R., Mittal, A., Kumar, M., & Mehta, P. K. (2016). Microbial proteases in commercial applications. J Pharm Chem Biol Sci, 4(3), 365–374.
Gupta, R., Gupta, K., Saxena, R. K., & Khan, S. (1999). Bleach-stable, alkaline protease from Bacillus sp. Biotechnology Letters, 21(2), 135–138.
Arya, P., Jani, S. A., Rajput, K. N., & Raval, V. (2020, March). Thermostable alkaline proteases from bacteria: A review. In Proceedings of the National Conference on Innovations in Biological Sciences (NCIBS).
Rawlings, N. D., & Barrett, A. J. (1993). Evolutionary families of peptidases. The Biochemical Journal, 290(1), 205–218.
Gurupriya, V. S., & Roy, S. C. (2017). In proteases in physiology and pathology. (Chakraborti, S., & Dhalla, N. S.).Springer, Singapore. pp. 195–216.
Sawant, R., & Nagendran, S. (2014). Protease: An enzyme with multiple industrial applications. World J Pharm Pharm Sci., 3(6), 568–579.
Sani, J. T., Gharibi, S. O. S., & Shariati, M. A. (2017). The importance of alkaline protease commercial applications: A short review. Indian J. Biotech. Pharm. Biotech., 5(1), 5.
Rao, M. B., Tanksale, A. M., Ghatge, M. S., & Deshpande, V. V. (1998). Molecular and biotechnological aspects of microbial proteases. Microbiology and Molecular Biology Reviews, 62(3), 597–635.
Mótyán, J. A., Tóth, F., & Tőzsér, J. (2013). Research applications of proteolytic enzymes in molecular biology. Biomol, 3(4), 923–942.
Dunaevsky, Y. E., Popova, V. V., Semenova, T. A., Beliakova, G. A., & Belozersky, M. A. (2014). Fungal inhibitors of proteolytic enzymes: Classification, properties, possible biological roles, and perspectives for practical use. Biochim., 101, 10–20.
Page, M. J., & Di Cera, E. (2008). Serine peptidases: Classification, structure and function. Cellular and Molecular Life Sciences, 65(7), 1220–1236.
Thakur, N., Goyal, M., Sharma, S., & Kumar, D. (2018). Proteases: Industrial applications and approaches used in strain improvement. In Biol Forum., 10(1), 158–167.
Moon, S. H., & Parulekar, S. J. (1991). A parametric study ot protease production in batch and fed-batch cultures of Bacillus firmus. Biotechnology and Bioengineering, 37(5), 467–483.
Rani, K., Rana, R., & Datt, S. (2012). Review on latest overview of proteases. Int J Curr Life Sci, 2(1), 12–18.
Badgujar, S. B., Mahajan, R. T., & Badgujar, S. B. (2010). Biological aspects of proteolytic enzymes: A review. J Pharm Res..
Fitzgerald, P. M., & Springer, J. P. (1991). Structure and function of retroviral proteases. Annual Review of Biophysics and Biophysical Chemistry, 20, 299–320.
Henry, J. P. (1992). Biological basis of the stress response. Integrative Psychological & Behavioral Science, 27(1), 66–83.
Szecsi, P. B. (1992). The aspartic proteases. Scandinavian Journal of Clinical and Laboratory Investigation, 52(210), 5–22.
Fricker, S. P. (2010). Cysteine proteases as targets for metal-based drugs. Metallomics, 2(6), 366–377.
Naveed, M., Nadeem, F., Mehmood, T., Bilal, M., Anwar, Z., & Amjad, F. (2021). Protease—A versatile and ecofriendly biocatalyst with multi-industrial applications: An updated review. Catal Letters, 151(2), 307–323.
Hashim, M. M., Mingsheng, D., Iqbal, M. F., & Xiaohong, C. (2011). Ginger rhizome as a potential source of milk coagulating cysteine protease. Phytochem., 72(6), 458–464.
Hashimoto, K., Yamanishi, Y., Maeyens, E., Dabbous, K., & Kanzaki, T. (1973). Collagenolytic activities of squamous cell carcinoma of the skin. Cancer Research, 33, 2790–2802.
Hibbs, M. S., Hasty, K. A., Seyer, J. M., Kang, A. H., & Mainardi, C. L. (1985). Biochemical and immunological characterization of the secreted forms of human neutrophil gelatinase. Journal of Biological Chemistry, 260(4), 2493–2500.
Chen, M. S., Lin, C. Y., Chiu, Y. H., Chen, C. P., Tsai, P. J., & Wang, H. S. (2018). IL-1β-Induced matrix metalloprotease-1 promotes mesenchymal stem cell migration via PAR1 and G-protein-coupled signaling pathway. Stem Cells Int., 2018, 3524759.
Ellaiah, P., Srinivasulu, B., & Adinarayana, K. (2002). A review on microbial alkaline proteases.
Banerjee, S., Maiti, T. K., & Roy, R. N. (2017). Protease production by thermo-alkaliphilic novel gut isolate Kitasatospora cheerisanensis GAP 124 from Gryllotalpa africana. Biocatal. Biotransformation, 35(3), 168–176.
de Castro, R. J. S., & Sato, H. H. (2014). Production and biochemical characterization of protease from Aspergillus oryzae: An evaluation of the physical–chemical parameters using agroindustrial wastes as supports. Biocatalysis and Agricultural Biotechnology, 3(3), 20–25.
Mishra, S. S., Ray, R. C., Rosell, C. M., & Panda, D. (2017). In microbial enzyme technology in food applications. CRC Press. US. pp. 3–18.
Tavano, O. L., Berenguer-Murcia, A., Secundo, F., & Fernandez-Lafuente, R. (2018). Biotechnological applications of proteases in food technology. Compr. Rev. Food Sci. Food Saf., 17(2), 412–436.
Fazilat, A. (2016). Production, isolation, purification and partial characterization of an extracellular acid protease from Aspergillus niger. Int J Adv Res Biol Sci, 3(3), 32–38.
Singh, S., & Bajaj, B. K. (2017). Potential application spectrum of microbial proteases for clean and green industrial production. Energy Ecol. Environ., 2(6), 370–386.
Contesini, F. J., Melo, R. R. D., & Sato, H. H. (2018). An overview of Bacillus proteases: From production to application. Critical Reviews in Biotechnology, 38(3), 321–334.
Jisha, V. N., Smitha, R. B., Pradeep, S., Sreedevi, S., Unni, K. N., Sajith, S., ... & Benjaminn, S. (2013). Versatility of microbial proteases Advances in Enzyme Research, 1 (3), 39–51.
Bhunia, B., Basak, B., & Dey, A. (2012). A review on production of serine alkaline protease by Bacillus spp. Journal of Biochemical Technology, 3(4), 448–457.
Chaud, L. C., Lario, L. D., Bonugli-Santos, R. C., Sette, L. D., Junior, A. P., & Felipe, M. D. G. D. A. (2016). Improvement in extracellular protease production by the marine antarctic yeast Rhodotorula mucilaginosa L7. New Biotechnology, 33(6), 807–814.
Jaswal, R. K., Kocher, G. S., & Virk, M. S. (2008). Production of alkaline protease by Bacillus circulans using agricultural residues: A statistical approach.
Al-Qodah, Z., Daghistani, H., & Alananbeh, K. (2013). Isolation and characterization of thermostable protease producing Bacillus pumilus from thermal spring in Jordan. Afr. J. Microbiol. Res., 7(29), 3711–3719.
Sharma, I., & Kango, N. (2021). Production and characterization of keratinase by Ochrobactrum intermedium for feather keratin utilization. International Journal of Biological Macromolecules, 166, 1046–1056.
Souza, P. M. D., Bittencourt, M. L. D. A., Caprara, C. C., Freitas, M. D., Almeida, R. P. C. D., Silveira, D., ... & Magalhães, P. O. (2015). A biotechnology perspective of fungal proteases. Braz. J. Microbiol., 46, 337-346.
Benluvankar, V., Jebapriya, G. R., & Gnanadoss, J. J. (2015). Protease production by Penicillium sp. LCJ228 under solid state fermentation using groundnut oilcake as substrate. Life., 50, 12.
Mukhtar, H., & Haq, I. (2013). Comparative evaluation of agroindustrial byproducts for the production of alkaline protease by wild and mutant strains of Bacillus subtilis in submerged and solid state fermentation. Sci. World J., 2013.
Novelli, P. K., Barros, M. M., & Fleuri, L. F. (2016). Novel inexpensive fungi proteases: Production by solid state fermentation and characterization. Food Chemistry, 198, 119–124.
Oyeleke, S. B., Egwim, E. C., & Auta, S. H. (2010). Screening of Aspergillus flavus and Aspergillus fumigatus strains for extracellular protease enzyme production. J. Microbiol. Antimicrob., 2(7), 83–87.
Sharma, A. K., Sharma, V., Saxena, J., Yadav, B., Alam, A., & Prakash, A. (2015). Isolation and screening of extracellular protease enzyme from bacterial and fungal isolates of soil. Int J Sci Res Environ Sci, 3(9), 0334–0340.
Erdönmez, O., Gedikli, S., Yaman, B. N., Çelik, P. A., & Çabuk, A. Thermophilic prokaryotic diversity of eynal geothermal spring and protease production potentials of isolates. Eskişehir. Tech. Univ. 21(3), 436-445.
Gurunathan, R., Huang, B., Ponnusamy, V. K., Hwang, J. S., & Dahms, H. U. (2021). Novel recombinant keratin degrading subtilisin like serine alkaline protease from Bacillus cereus isolated from marine hydrothermal vent crabs. Science and Reports, 11(1), 1–14.
Akkır, E. Y., Şahin, Y. B., Gedikli, S., Çelik, P. A., & Çabuk, A. (2021). Extremely thermostable, EDTA-resistant alkaline protease from a thermophilic Geobacillus subterraneus C2–1 isolate. J. microbiol., biotechnol. food sci. 2021, 50–56.
Emran, M. A., Ismail, S. A., & Hashem, A. M. (2020). Production of detergent stable thermophilic alkaline protease by Bacillus licheniformis ALW1. Biocatal. Agric. Biotechnol., 26, 101631.
Arya, P., Trivedi, M., Ramani, N., Patel, K., & Rajput, K. N. (2020). Isolation, production and applications of alkaline protease from hot springs bacterial isolates. Biosci. Biotechnol. Res. Commun..
Naidu, K. S. B., & Devi, K. L. (2005). Optimization of thermostable alkaline protease production from species of Bacillus using rice bran. African Journal of Biotechnology, 4(7), 724–726.
Ginting, E. L., Kemer, K., Wullur, S., & Uria, A. R. (2020). Identification of proteolytic thermophiles from Moinit Coastal hot-spring, North Sulawesi. Indonesia. Geomicrobiol. J., 37(1), 50–58.
Mechri, S., Bouacem, K., Jabeur, F., Mohamed, S., Addou, N. A., & Dab, A.,... & Jaouadi, B. . (2019). Purification and biochemical characterization of a novel thermostable and halotolerant subtilisin SAPN, a serine protease from Melghiribacillus thermohalophilus Nari2A T for chitin extraction from crab and shrimp shell by-products. Extremophiles, 23(5), 529–547.
Manavalan, T., Manavalan, A., Ramachandran, S., & Heese, K. (2020). Identification of a novel thermostable alkaline protease from Bacillus megaterium-TK1 for the detergent and leather industry. Biology., 9(12), 472.
Nnolim, N. E., Okoh, A. I., & Nwodo, U. U. (2020). Proteolytic bacteria isolated from agro-waste dumpsites produced keratinolytic enzymes. Biotechnol. Rep., 27, e00483.
Lam, M. Q., Mut, N. N. N., Thevarajoo, S., Chen, S. J., Selvaratnam, C., Hussin, H., ... & Chong, C. S. (2018). Characterization of detergent compatible protease from halophilic Virgibacillus sp. CD6. 3 Biotech., 8(2), 1–9.
Kaewsalud, T., Yakul, K., Jantanasakulwong, K., Tapingkae, W., Watanabe, M., & Chaiyaso, T. (2020). Biochemical characterization and application of thermostable-alkaline keratinase from Bacillus halodurans SW-X to valorize chicken feather wastes. Waste and Biomass Valorization, 1–14.
Tuysuz, E., Gonul-Baltaci, N., Omeroglu, M. A., Adiguzel, A., Taskin, M., & Ozkan, H. (2020). Co-production of amylase and protease by locally isolated thermophilic bacterium Anoxybacillus rupiensis T2 in sterile and non-sterile media using waste potato peels as substrate. Waste and Biomass Valorization, 1–10.
Nnolim, N. E., & Nwodo, U. U. (2020). Bacillus sp. CSK2 produced thermostable alkaline keratinase using agro-wastes: Keratinolytic enzyme characterization. BMC Biotechnol., 20(1), 65.
Vidyasagar, M., Prakash, S. B., & Sreeramulu, K. (2006). Optimization of culture conditions for the production of haloalkaliphilic thermostable protease from an extremely halophilic archaeon Halogeometricum sp. TSS101. Lett. Appl. Microbiol., 43(4), 385–391.
Sharma, A. K., Kikani, B. A., & Singh, S. P. (2020). Biochemical, thermodynamic and structural characteristics of a biotechnologically compatible alkaline protease from a haloalkaliphilic, Nocardiopsis dassonvillei OK-18. International Journal of Biological Macromolecules, 153, 680–696.
Tsuchiya, K., Nakamura, Y., Sakashita, H., & Kimura, T. (1992). Purification and characterization of a thermostable alkaline protease from alkalophilic Thermoactinomyces sp. HS682. Biosci. Biotechnol. Biochem., 56(2), 246–250.
Tsuchiya, K., Arai, T., Seki, K., & Kimura, T. (1987). Purification and some properties of alkaline proteinases from Cephalosporium sp. KM388. Agric. Biol. Chem., 51(11), 2959–2965.
Damare, S., Mishra, A., D’Souza-Ticlo-Diniz, D., Krishnaswamy, A., & Raghukumar, C. (2020). A deep-sea hydrogen peroxide-stable alkaline serine protease from Aspergillus flavus. 3 Biotech, 10(12), 1–9.
Abu-Tahon, M. A., Arafat, H. H., & Isaac, G. S. (2020). Laundry detergent compatibility and dehairing efficiency of alkaline thermostable protease produced from Aspergillus terreus under solid-state fermentation. Journal of Oleo Science, 69(3), 241–254.
Ja’afaru, M. I., Chimbekujwo, K. I., & Ajunwa, O. M. . (2020). Purification, characterization and de-staining potentials of a thermotolerant protease produced by Fusarium oxysporum. Period. Polytech. Chem. Eng., 64(4), 539–547.
Nieri, B., Canino, S., Versace, R., & Alpi, A. (1998). Purification and characterization of an endoprotease from alfalfa senescent leaves. Phytochemistry, 49(3), 643–649.
Hölker, U., Höfer, M., & Lenz, J. (2004). Biotechnological advantages of laboratory-scale solid-state fermentation with fungi. Applied Microbiology and Biotechnology, 64(2), 175–186.
Silva, B. L., Geraldes, F. M., Murari, C. S., Gomes, E., & Da-Silva, R. (2014). Production and characterization of a milk-clotting protease produced in submerged fermentation by the thermophilic fungus Thermomucor indicae-seudaticae N31. Applied Biochemistry and Biotechnology, 172(4), 1999–2011.
Bonugli-Santos, R. C., dos Santos Vasconcelos, M. R., Passarini, M. R., Vieira, G. A., Lopes, V. C., Mainardi, P. H., ... & Sette, L. D. (2015). Marine-derived fungi: Diversity of enzymes and biotechnological applications. Front. Microbiol., 6, 269.
Kasana, R. C., Salwan, R., & Yadav, S. K. (2011). Microbial proteases: Detection, production, and genetic improvement. Critical Reviews in Microbiology, 37(3), 262–276.
Lemes, A. C., Álvares, G. T., Egea, M. B., Brandelli, A., & Kalil, S. J. (2016). Simultaneous production of proteases and antioxidant compounds from agro-industrial by-products. Bioresource Technology, 222, 210–216.
Rathod, M. G., & Pathak, A. P. (2014). Wealth from waste: Optimized alkaline protease production from agro-industrial residues by Bacillus alcalophilus LW8 and its biotechnological applications. J. Taibah Univ. Sci., 8(4), 307–314.
Subramaniyam, R., & Vimala, R. (2012). Solid state and submerged fermentation for the production of bioactive substances: A comparative study. Int J Sci Nat, 3(3), 480–486.
Simair, A. A., Qureshi, A. S., Khushk, I., Ali, C. H., Lashari, S., Bhutto, M. A., ... & Lu, C. (2017). Production and partial characterization of α-amylase enzyme from Bacillus sp. BCC 01–50 and potential applications. Biomed Res. Int., 2017.
Chutmanop, J., Chuichulcherm, S., Chisti, Y., & Srinophakun, P. (2008). Protease production by Aspergillus oryzae in solid-state fermentation using agroindustrial substrates. Journal of Chemical Technology and Biotechnology, 83(7), 1012–1018.
Niyonzima, F. N., & More, S. S. (2013). Screening and optimization of cultural parameters for an alkaline protease production by Aspergillus terreus gr. under submerged fermentation. Int J Pharm Bio Sci, 4(1), 1016–1028.
Kumar, A. G., Swarnalatha, S., Gayathri, S., Nagesh, N., & Sekaran, G. (2008). Characterization of an alkaline active–thiol forming extracellular serine keratinase by the newly isolated Bacillus pumilus. Journal of Applied Microbiology, 104(2), 411–419.
Lee, H., Suh, D. B., Hwang, J. H., & Suh, H. J. (2002). Characterization of a kerationlytic metalloprotease from Bacillus sp. SCB-3. Appl. Biochem. Biotechnol., 97(2), 123–133.
Mitsuiki, S., Sakai, M., Moriyama, Y., Goto, M., & Furukawa, K. (2002). Purification and some properties of a keratinolytic enzyme from an alkaliphilic Nocardiopsis sp. TOA-1. Biosci. Biotechnol. Biochem., 66(1), 164–167.
Nam, G. W., Lee, D. W., Lee, H. S., Lee, N. J., Kim, B. C., Choe, E. A., ... & Pyun, Y. R. (2002). Native-feather degradation by Fervidobacterium islandicum AW-1, a newly isolated keratinase-producing thermophilic anaerobe. Arch. Microbiol., 178(6), 538-547.
Cao, Z. J., Zhang, Q., Wei, D. K., Chen, L., Wang, J., Zhang, X. Q., & Zhou, M. H. (2009). Characterization of a novel Stenotrophomonas isolate with high keratinase activity and purification of the enzyme. Journal of Industrial Microbiology, 36(2), 181–188.
Benito, M. J., Rodríguez, M., Núnez, F., Asensio, M. A., Bermúdez, M. E., & Córdoba, J. J. (2002). Purification and characterization of an extracellular protease from Penicillium chrysogenum Pg222 active against meat proteins. Applied and Environment Microbiology, 68(7), 3532–3536.
Damare, S., Raghukumar, C., Muraleedharan, U. D., & Raghukumar, S. (2006). Deep-sea fungi as a source of alkaline and cold-tolerant proteases. Enyzme and Microbial Technology, 39(2), 172–181.
Devi, M. K., Banu, A. R., Gnanaprabhal, G. R., Pradeep, B. V., & Palaniswamy, M. (2008). Purification, characterization of alkaline protease enzyme from native isolate Aspergillus niger and its compatibility with commercial detergents. Indian Journal of Science and Technology, 1(7), 1–6.
Ali, T. H., Ali, N. H., & Mohamed, L. A. (2011). Production, purification and some properties of extracellular keratinase from feathers-degradation by Aspergillus oryzae Nrrl-447. J. Appl. Sci. Environ., 6(2).
Razzaq, A., Shamsi, S., Ali, A., Ali, Q., Sajjad, M., Malik, A., & Ashraf, M. (2019). Microbial proteases applications. Front. Bioeng. Biotechnol., 7, 110.
Sharma, M., Gat, Y., Arya, S., Kumar, V., Panghal, A., & Kumar, A. (2019). A review on microbial alkaline protease: An essential tool for various industrial approaches. Industrial Biotechnology, 15(2), 69–78.
Prakash, O., Nimonkar, Y., Chavadar, M. S., Bharti, N., Pawar, S., Sharma, A., & Shouche, Y. S. (2017). Optimization of nutrients and culture conditions for alkaline protease production using two endophytic Micrococci: Micrococcus aloeverae and Micrococcus yunnanensis. Indian J. Microbiol., 57(2), 218–225.
Qureshi, A. S., Simair, A. A., Ali, C. H., Khushk, I., Khokhar, J. A., Ahmad, A., & Lu, C. (2018). Production, purification and partial characterization of organo-solvent tolerant protease from newly isolated Bacillus sp. BBXS-2. Ferment Technol, 7(151), 2.
Malathi, S., Priya, D. M., & Palani, P. (2014). Microbial diversity and biotechnology in food security. (Kharwar, R. N., Upadhyay, R. S., Dubey, N. K., & Raghuwanshi), R. Springer, New Delhi. pp. 451–462.
Shanmuga Priya, S., Krishnareni, J., Selvin, J., Gandhimathi, R., Arunkumar, M., Thangavelu, T., ... & Natarajaseenivasan, K. (2008). Optimization of extracellular thermotolerant alkaline protease produced by marine Roseobacter Sp (MMD040). Bioprocess Biosyst Eng, 31, 427-433.
Abd Rahman, R. N. Z., Geok, L. P., Basri, M., & Salleh, A. B. (2005). Physical factors affecting the production of organic solvent-tolerant protease by Pseudomonas aeruginosa strain K. Bioresource Technology, 96(4), 429–436.
Mothe, T., & Sultanpuram, V. R. (2016). Production, purification and characterization of a thermotolerant alkaline serine protease from a novel species Bacillus caseinilyticus. 3 Biotech, 6(1), 53.
Pandey, A., Tyagi, R. D., & Varjani, S. (Eds.). (2021). Biomass, biofuels, biochemicals: Circular bioeconomy—Current developments and future outlook. Elsevier.
Wilson, P., & Remigio, Z. (2012). Production and characterisation of protease enzyme produced by a novel moderate thermophilic bacterium (EP1001) isolated from an alkaline hot spring. Zimbabwe. Afr. J. Microbiol. Res., 6(27), 5542–5551.
Govind, N. S., Mehta, B., Sharma, M., & Modi, V. V. (1981). Protease and carotenogenesis
Deutscher, M. P. (Ed.). (1990). Guide to protein purification. Gulf Professional Publishing.
Lakshmi, B. K. M., Kumar, D. M., & Hemalatha, K. P. J. (2018). Purification and characterization of alkaline protease with novel properties from Bacillus cereus strain S8. Journal, Genetic Engineering & Biotechnology, 16(2), 295–304.
Banerjee, G., & Ray, A. K. (2017). Impact of microbial proteases on biotechnological industries. Biotechnology and Genetic Engineering Reviews, 33(2), 119–143.
Kumar, C. G., & Takagi, H. (1999). Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnology Advances, 17(7), 561–594.
Iqbal, M., Tao, Y., Xie, S., Zhu, Y., Chen, D., & Wang, X.,... & Yuan, Z. . (2016). Aqueous two-phase system (ATPS): An overview and advances in its applications. Biol. Proced. Online, 18(1), 1–18.
Hotha, S., & Banik, R. M. (1997). Production of alkaline protease by Bacillus thuringiensis H 14 in Aqueous Two-Phase Systems. Int. J. Environ. Res, 69(1), 5–10.
Jani, S. A., Heta, R., Patel, H., Darji, D., Rathod, A., & Pal, S. (2015). Production and characterization of keratinolytic protease from Streptomyces sp. International Journal of Current Microbiology and Applied Sciences, 4(5), 962–975.
Hajji, M., Rebai, A., Gharsallah, N., & Nasri, M. (2008). Optimization of alkaline protease production by Aspergillus clavatus ES1 in Mirabilis jalapa tuber powder using statistical experimental design. Applied Microbiology and Biotechnology, 79(6), 915–923.
Joo, H. S., Kumar, C. G., Park, G. C., Kim, K. T., Paik, S. R., & Chang, C. S. (2002). Optimization of the production of an extracellular alkaline protease from Bacillus horikoshii. Process Biochemistry, 38(2), 155–159.
Watanabe, A., Kamio, Y., Kimura, W., & Izaki, K. (1993). Purification and characterization of a thermostable neutral metalloprotease I from Chloroflexus aurantiacus J-10-fl. Bioscience, Biotechnology, and Biochemistry, 57(12), 2160–2165.
Adinarayana, K., Ellaiah, P., & Prasad, D. S. (2003). Purification and partial characterization of thermostable serine alkaline protease from a newly isolated Bacillus subtilis PE-11. An Official Journal of the American Association of Pharmaceutical Scientists, 4(4), 440–448.
Asker, M. M., Mahmoud, M. G., El Shebwy, K., & Abd el Aziz, M. S. (2013). Purification and characterization of two thermostable protease fractions from Bacillus megaterium. Journal, Genetic Engineering & Biotechnology, 11(2), 103–109.
Pathak, A. P., & Sardar, A. G. (2014). Isolation and characterization of salt stable protease producing archaea from marine solar saltern of Mulund, Mumbai.
Yilmaz, B., Baltaci, M. O., Sisecioglu, M., & Adiguzel, A. (2016). Thermotolerant alkaline protease enzyme from Bacillus licheniformis A10: Purification, characterization, effects of surfactants and organic solvents. Journal of Enzyme Inhibition and Medicinal Chemistry, 31(6), 1241–1247.
Phadatare, S. U., Deshpande, V. V., & Srinivasan, M. C. (1993). High activity alkaline protease from Conidiobolus coronatus (NCL 86.8. 20): Enzyme production and compatibility with commercial detergents. Enzyme Microb. Technol., 15(1), 72–76.
Gupta, R., & Ramnani, P. (2006). Microbial keratinases and their prospective applications: An overview. Appl. Microbiol. Biotechno, 70(1), 21–33.
Pandey, S., Rakholiya, K. D., Raval, V. H., & Singh, S. P. (2012). Catalysis and stability of an alkaline protease from a haloalkaliphilic bacterium under non-aqueous conditions as a function of pH, salt and temperature. Journal of Bioscience and Bioengineering, 114(3), 251–256.
Tsai, Y. C., Juang, R. Y., Lin, S. F., Chen, S. W., Yamasaki, M., & Tamura, G. (1988). Production and further characterization of an alkaline elastase produced by alkalophilic Bacillus strain Ya-B. Applied and Environment Microbiology, 54(12), 3156–3161.
Huston, A. L., Methe, B., & Deming, J. W. (2004). Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Applied and Environment Microbiology, 70(6), 3321–3328.
Verma, J., & Pandey, S. (2019). Characterization of partially purified alkaline protease secreted by halophilic bacterium Citricoccus sp. isolated from agricultural soil of northern India. Biocatalysis and Agricultural Biotechnology, 17, 605–612.
Koszelak, S., Ng, J. D., Day, J., Ko, T. P., Greenwood, A., & McPherson, A. (1997). The crystallographic structure of the subtilisin protease from Penicillium cyclopium. Biochem., 36(22), 6597–6604.
Dadshahi, Z., Homaei, A., Zeinali, F., Sajedi, R. H., & Khajeh, K. (2016). Extraction and purification of a highly thermostable alkaline caseinolytic protease from wastes Penaeus vannamei suitable for food and detergent industries. Food Chemistry, 202, 110–115.
Silva, C., Martins, M., Jing, S., Fu, J., & Cavaco-Paulo, A. (2018). Practical insights on enzyme stabilization. Critical Reviews in Biotechnology, 38(3), 335–350.
Xia, W., Xu, X., Qian, L., Shi, P., Bai, Y., Luo, H., ... & Yao, B. (2016). Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharification performance. Biotechnol. Biofuels, 9(1), 1-12.
Hasunuma, T., Okazaki, F., Okai, N., Hara, K. Y., Ishii, J., & Kondo, A. (2013). A review of enzymes and microbes for lignocellulosic biorefinery and the possibility of their application to consolidated bioprocessing technology. Bioresource Technology, 135, 513–522.
Bilal, M., Zhao, Y., Noreen, S., Shah, S. Z. H., Bharagava, R. N., & Iqbal, H. M. (2019). Modifying bio-catalytic properties of enzymes for efficient biocatalysis: A review from immobilization strategies viewpoint. Biocatal. Biotransformation, 37(3), 159–182.
Wu, I., & Arnold, F. H. (2013). Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures. Biotechnology and Bioengineering, 110(7), 1874–1883.
Woodley, J. M. (2018). Integrating protein engineering with process design for biocatalysis. A Math Phys Eng Sci., 376(2110), 20170062.
Wang, Q., Zhou, L., Jiang, Y., & Gao, J. (2011). Improved stability of the carbon nanotubes–enzyme bioconjugates by biomimetic silicification. Enyzme and Microbial Technology, 49(1), 11–16.
Blum, J. K., Ricketts, M. D., & Bommarius, A. S. (2012). Improved thermostability of AEH by combining B-FIT analysis and structure-guided consensus method. Journal of Biotechnology, 160(3–4), 214–221.
Pickersgill, R. W., Sumner, I. G., & Goodenough, P. W. (1990). Preliminary crystallographic data for protease. European Journal of Biochemistry, 190(2), 443–444.
Ashraf, N. M., Krishnagopal, A., Hussain, A., Kastner, D., Sayed, A. M. M., Mok, Y. K., & Zeeshan, N. (2019). Engineering of serine protease for improved thermostability and catalytic activity using rational design. International Journal of Biological Macromolecules, 126, 229–237.
Li, Y., Hu, F., Wang, X., Cao, H., Liu, D., & Yao, D. (2013). A rational design for trypsin-resistant improvement of Armillariella tabescens β-mannanase MAN47 based on molecular structure evaluation. Journal of Biotechnology, 163(4), 401–407.
Xu, M. Q., Wang, S. S., Li, L. N., Gao, J., & Zhang, Y. W. (2018). Combined cross-linked enzyme aggregates as. Biocatal., 8(10), 460.
Hu, W., Liu, X., Li, Y., Liu, D., Kuang, Z., Qian, C., & Yao, D. (2017). Rational design for the stability improvement of Armillariella tabescens β-mannanase MAN47 based on N-glycosylation modification. Enyzme and Microbial Technology, 97, 82–89.
Reetz, M. T., & Carballeira, J. D. (2007). Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat., 2(4), 891–903.
Porter, J. L., Rusli, R. A., & Ollis, D. L. (2016). Directed evolution of enzymes for industrial biocatalysis. Chem Bio Chem, 17(3), 197–203.
Zhu, F., He, B., Gu, F., Deng, H., Chen, C., Wang, W., & Chen, N. (2020). Improvement in organic solvent resistance and activity of metalloprotease by directed evolution. Journal of Biotechnology, 309, 68–74.
Zhao, H. Y., Wu, L. Y., Liu, G., & Feng, H. (2016). Single-site substitutions improve cold activity and increase thermostability of the dehairing alkaline protease (DHAP). Bioscience, Biotechnology, and Biochemistry, 80(12), 2480–2485.
Fang, Z., Zhang, J., Du, G., & Chen, J. (2016). Improved catalytic efficiency, thermophilicity, anti-salt and detergent tolerance of keratinase KerSMD by partially truncation of PPC domain. Science and Reports, 6(1), 1–11.
Guo, Y., Tu, T., Zheng, J., Ren, Y., Wang, Y., Bai, Y.,... & Luo, H. (2020). A novel thermostable aspartic protease from Talaromyces leycettanus and its specific autocatalytic activation through an intermediate transition state. Appl. Microbiol. Biotechnol., 104(11), 4915-4926.
Morsi, R., Bilal, M., Iqbal, H. M., & Ashraf, S. S. (2020). Laccases and peroxidases: the smart, greener and futuristic biocatalytic tools to mitigate recalcitrant emerging pollutants. Sci. Total Environ., 714, 136572.
Bilal, M., & Iqbal, H. M. (2019). Chemical, physical, and biological coordination: An interplay between materials and enzymes as potential platforms for immobilization. Coordination Chemistry Reviews, 388, 1–23.
Tanksale, A., Chandra, P. M., Rao, M., & Deshpande, V. (2001). Immobilization of alkaline protease from Conidiobolus macrosporus for reuse and improved thermal stability. Journal of Biotechnology Letters, 23(1), 51–54.
Ibrahim, A. S. S., Al-Salamah, A. A., El-Toni, A. M., Almaary, K. S., El-Tayeb, M. A., Elbadawi, Y. B., & Antranikian, G. (2016). Enhancement of alkaline protease activity and stability via covalent immobilization onto hollow core-mesoporous shell silica nanospheres. International Journal of Molecular Sciences, 17(2), 184.
Adamczak, M., & Krishna, S. H. (2004). Strategies for improving enzymes for efficient biocatalysis. Food Technol. Biotechnol., 42(4), 251–264.
Kumar, D., & Bhalla, T. C. (2005). Microbial proteases in peptide synthesis: Approaches and applications. Applied Microbiology and Biotechnology, 68(6), 726–736.
Awad, G. E., Ghanem, A. F., Wahab, W. A. A., & Wahba, M. I. (2020). Functionalized κ-carrageenan/hyperbranched poly (amidoamine) for protease immobilization: Thermodynamics and stability studies. International Journal of Biological Macromolecules, 148, 1140–1155.
Özacar, M., Mehde, A. A., Mehdi, W. A., Özacar, Z. Z., & Severgün, O. (2019). The novel multi cross-linked enzyme aggregates of protease, lipase, and catalase production from the sunflower seeds, characterization and application. Colloids Surf., B. 173, 58-68.
Bilal, M., Jing, Z., Zhao, Y., & Iqbal, H. M. (2019). Immobilization of fungal laccase on glutaraldehyde cross-linked chitosan beads and its bio-catalytic potential to degrade bisphenol A. Biocatal. Agric. Biotechnol., 19, 101174.
de Melo Brites, M., Cerón, A. A., Costa, S. M., Oliveira, R. C., Ferraz, H. G., Catalani, L. H., & Costa, S. A. (2020). Bromelain immobilization in cellulose triacetate nanofiber membranes from sugarcane bagasse by electrospinning technique. Enzyme Microb. Technol., 132, 109384.
Benucci, I., Caso, M. C., Bavaro, T., Masci, S., Keršienė, M., & Esti, M. (2020). Prolyl endopeptidase from Aspergillus niger immobilized on a food-grade carrier for the production of gluten-reduced beer. Food Control, 110, 106987.
Hu, T. G., Cheng, J. H., Zhang, B. B., Lou, W. Y., & Zong, M. H. (2015). Immobilization of alkaline protease on amino-functionalized magnetic nanoparticles and its efficient use for preparation of oat polypeptides. Industrial and Engineering Chemistry Research, 54(17), 4689–4698.
Wang, S. N., Zhang, C. R., Qi, B. K., Sui, X. N., Jiang, L. Z., Li, Y., & Zhang, Q. Z. (2014). Immobilized alcalase alkaline protease on the magnetic chitosan nanoparticles used for soy protein isolate hydrolysis. European Food Research and Technology, 239(6), 1051–1059.
Thakrar, F. J., & Singh, S. P. (2019). Catalytic, thermodynamic and structural properties of an immobilized and highly thermostable alkaline protease from a haloalkaliphilic actinobacteria, Nocardiopsis alba TATA-5. Bioresource Technology, 278, 150–158.
Sadjadi, M. S., Farhadyar, N., & Zare, K. (2009). Synthesis of bi-metallic Au–Ag nanoparticles loaded on functionalized MCM-41 for immobilization of alkaline protease and study of its biocatalytic activity. Superlattices and Microstructures, 46(4), 563–571.
Icimoto, M. Y., Brito, A. M. M., Ramos, M. P. C., Oliveira, V., & Nantes-Cardoso, I. L. (2020). Increased stability of oligopeptidases immobilized on gold nanoparticles. Catalysts, 10(1), 78.
Sahu, A., Badhe, P. S., Adivarekar, R., Ladole, M. R., & Pandit, A. B. (2016). Synthesis of glycinamides using protease immobilized magnetic nanoparticles. Biotechnol. Rep, 12, 13–25.
Panda, S. K., Mishra, S. S., Kayitesi, E., & Ray, R. C. (2016). Microbial-processing of fruit and vegetable wastes for production of vital enzymes and organic acids: Biotechnology and scopes. Environmental Research, 146, 161–172.
Leisola, M., Jokela, J., Pastinen, O., Turunen, O., & Schoemaker, H. (2001). Industrial use of enzymes. Eolss Publishers.
Anwar, A., & Saleemuddin, M. (1998). Alkaline proteases: A review. Bioresource Technology, 64(3), 175–183.
Samal, B. B., Karan, B., & Stabinsky, Y. (1990). Stability of two novel serine proteinases in commercial laundry detergent formulations. Biotechnology and Bioengineering, 35(6), 650–652.
Yadav, S. K., Bisht, D., Shikha, S., & Darmwal, N. S. (2011). Oxidant and solvent stable alkaline protease from Aspergillus flavus and its characterization. African Journal of Biotechnology, 10(43), 8630–8640.
Rao, K., & Narasu, M. L. (2007). Alkaline protease from Bacillus firmus 7728. Afr. j. biotechnol., 6(21).
Benmrad, M. O., Moujehed, E., Elhoul, M. B., Mechri, S., Bejar, S., Zouari, R., ... & Jaouadi, B. (2018). Production, purification, and biochemical characterization of serine alkaline protease from Penicillium chrysogenium strain X5 used as excellent bio-additive for textile processing. Int. J. Biol. Macromol., 119, 1002-1016.
Wahab, W. A. A., & Ahmed, S. A. (2018). Response surface methodology for production, characterization and application of solvent, salt and alkali-tolerant alkaline protease from isolated fungal strain Aspergillus niger WA 2017. International Journal of Biological Macromolecules, 115, 447–458.
Dettmer, A., Cavalli, É., Ayub, M. A., & Gutterres, M. (2013). Environmentally friendly hide unhairing: Enzymatic hide processing for the replacement of sodium sulfide and delimig. Journal of Cleaner Production, 47, 11–18.
Dayanandan, A., Kanagaraj, J., Sounderraj, L., Govindaraju, R., & Rajkumar, G. S. (2003). Application of an alkaline protease in leather processing: An ecofriendly approach. Journal of Cleaner Production, 11(5), 533–536.
Ahmed, S. A., Al-domany, R. A., El-Shayeb, N. M., Radwan, H. H., & Saleh, S. A. (2008). Optimization, immobilization of extracellular alkaline protease and characterization of its enzymatic properties. Research Journal of Agriculture and Biological Sciences, 4(5), 434–446.
Kainoor, P. S., & Naik, G. R. (2010). Production and characterization of feather degrading keratinase from Bacillus sp. JB 99.
Verma, A., Singh, H., Anwar, S., Chattopadhyay, A., Tiwari, K. K., Kaur, S., & Dhilon, G. S. (2017). Microbial keratinases: Industrial enzymes with waste management potential. Critical Reviews in Biotechnology, 37(4), 476–491.
Tapia, D. M., & Simões, M. L. G. (2008). Production and partial characterization of keratinase produced by a microorganism isolated from poultry processing plant wastewater. Afr. j. biotechnol., 7(3).
Dominguez-Benetton, X., Srikanth, S., Satyawali, Y., Vanbroekhoven, K., & Pant, D. (2013). Enzymatic electrosynthesis: An overview on the progress in enzyme-electrodes for the production of electricity, fuels and chemicals. J Microb Biochem Technol, 6, 007.
Donato, R. K., & Mija, A. (2020). Keratin associations with synthetic, biosynthetic and natural polymers: An extensive review. Polymers, 12(1), 32.
Sionkowska, A., Kaczmarek, B., Michalska, M., Lewandowska, K., & Grabska, S. (2017). Preparation and characterization of collagen/chitosan/hyaluronic acid thin films for application in hair care cosmetics. Pure and Applied Chemistry, 89(12), 1829–1839.
Kojima, M., Kanai, M., Tominaga, M., Kitazume, S., Inoue, A., & Horikoshi, K. (2006). Isolation and characterization of a feather-degrading enzyme from Bacillus pseudofirmus FA30-01. Extremophiles, 10(3), 229–235.
Tork, S., Aly, M. M., & Nawar, L. (2010). Biochemical and molecular characterization of a new local keratinase producing Pseudomomanas sp., MS21. Asian J Biotechnol, 2(1), 1–13.
Singhal, P., Nigam, V. K., & Vidyarthi, A. S. (2012). Studies on production, characterization and applications of microbial alkaline proteases. International Journal of Advanced Biotechnology and Research, 3(3), 653–669.
Raval, V. H., Purohit, M. K., & Singh, S. P. (2015). In Halophiles. (DK Maheshwari, M Saraf) Springer, Cham. pp. 421–449.
Guo, C., Li, C., & Kaplan, D. L. (2020). Enzymatic degradation of Bombyx mori silk materials: A review. Biomacromolecules, 21(5), 1678–1686.
Romsomsa, N., Chim-anagae, P., & Jangchud, A. (2010). Optimization of silk degumming protease production from Bacillus subtilis C4 using Plackett-Burman design and response surface methodology. Sci Asia, 36, 118–124.
Chauhan, B., & Gupta, R. (2004). Application of statistical experimental design for optimization of alkaline protease production from Bacillus sp. RGR-14. Process Biochem, 39(12), 2115–2122.
Chanalia, P., Gandhi, D., Jodha, D., & Singh, J. (2011). Applications of microbial proteases in pharmaceutical industry: An overview. Rev Med Microbiol, 22(4), 96–101.
Acknowledgements
Authors P. A. and R. V. kindly acknowledge the financial assistance and research grants under the “SHODH – Scheme of Developing High-Quality Research” fellowship by Department of Higher Education, Government of Gujarat Ref No: 201901380034 and UGC-BSR Start-Up Research Grant No.F.30-521/2020 by University Grants Commission, Government of India. The infrastructural support at the Department of Microbiology & Biotechnology, Gujarat University, Ahmedabad, is heartily acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors have read the manuscript and gave their consent for publication.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Arya, P.S., Yagnik, S.M., Rajput, K.N. et al. Understanding the Basis of Occurrence, Biosynthesis, and Implications of Thermostable Alkaline Proteases. Appl Biochem Biotechnol 193, 4113–4150 (2021). https://doi.org/10.1007/s12010-021-03701-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03701-x