Abstract
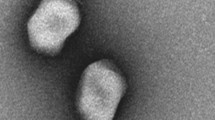
The genome sequence, morphology, and genetic features of a novel phage, named SSE1, is reported here. Phage SSE1 that infects Shigella dysenteriae (China General Microbiological Culture Collection Center number: 1.1869) was isolated from the aeration tank water of a sewage treatment plant. SSE1 showed morphological features associated with those of phages in Myoviridae. The whole genome sequence of phage SSE1 is composed of 169,744 bp with the GC content of 37.51%. The double-stranded DNA of SSE1 contains 270 open reading frameworks (ORFs). Phylogenetically, phage SSE1 showed a stronger homology (whole genome and terminase large subunit protein sequence) to Escherichia phages than other Shigella phages in the NCBI database, but SSE1 did not infect Escherichia stains. This indicates that phage SSE1 should be a novel phage infecting Shigella dysenteriae. Besides, the result of this study provided a new idea for phage therapy. SSE1 may become a candidate for potential therapy against Shigella dysenteriae infection in clinical applications.




Similar content being viewed by others
Availability of Data and Materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- S. dysenteriae :
-
Shigella dysenteriae
- CGMCC:
-
China General Microbiological Culture Collection Center
- ORFs:
-
Open reading frameworks
- LEDS:
-
Laboratory-based Enteric Disease Surveillance
- NB:
-
Nutrient Broth
- TEM:
-
Transmission electron microscopy
- EDTA:
-
Ethylene diamine tetraacetic acid
- SDS:
-
Sodium dodecyl sulfate
- E. coli :
-
Escherichia coli
- LB:
-
Luria-Bertani
- CDC:
-
Chinese Center for Disease Control and Prevention
- NCBI:
-
National Center for Biotechnology Information
- ICTV:
-
International Committee on Taxonomy of Viruses
References
Levine, M. M., DuPont, H. L., Formal, S. B., Hornick, R. B., Takeuchi, A., Gangarosa, E. J., Snyder, M. J., & Libonati, J. P. (1973). Pathogenesis of Shigella dysenteriae 1 (Shiga) dysentery. The Journal of Infectious Diseases, 127(3), 261–270.
Tóth, I., Sváb, D., Bálint, B., Brown-Jaque, M., & Maróti, G. (2016). Comparative analysis of the Shiga toxin converting bacteriophage first detected in Shigella sonnei. Infection, Genetics and Evolution, 37, 150–157.
Topka, G., Bloch, S., & Boz’ena Nejman-Falen’czyk, etc. (2018). Characterization of bacteriophage vB-EcoS-95, isolated from urban sewage and revealing extremely rapid lytic development. Frontiers in Microbiology, 9, 3326.
Davis, C. L., Wahid, R., Toapanta, F. R., Simon, J. K., & Sztein, M. B. (2018). A clinically parameterized mathematical model of Shigella immunity to inform vaccine design. PLoS One, 13(1), e0189571.
Terry, L. M., Barker, C. R., Day, M. R., Greig, D. R., Dallman, T. J., & Jenkins, C. (2018). Antimicrobial resistance profiles of Shigella dysenteriae isolated from travellers returning to the UK, 2004-2017. Journal of Medical Microbiology, 67(8), 1022–1030.
Centers for Disease Control and Prevention. National enteric disease surveillance: shigella annual report, 2016. https://www.cdc.gov/nationalsurveillance/pdfs/LEDS-Shig-2016-REPORT-508.pdf.
Centers for Disease Control and Prevention. Travel-related infectious diseases: shigellosis. https://wwwnc.cdc.gov/travel/yellowbook/2020/travelrelated-infectious-diseases/shigellosis.
World Health Organization. Disease Commodity Packages – Shigellosis. https://www.who.int/internal-publications-detail/shigellosis.
Abedon S. T. (2017). Bacteriophage clinical use as antibacterial “drugs”: Utility and precedent. Microbiology Spectrum, 5(4), BAD-0003-2016.
Centers for Disease Control and Prevention. Antibiotic/antimicrobial resistance. https://www.cdc.gov/drugresistance/.
Rohde, C., Resch, G., Pirnay, J. P., Blasdel, B. G., Debarbieux, L., Gelman, D., et al. (2018). Expert opinion on three phage therapy related topics: Bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses., 10(4), e178.
Mertz, L. (2019). Battling superbugs: How phage therapy went from obscure to promising. IEEE Pulse, 10(1), 3–9.
Dedrick, R. M., Guerrero-Bustamante, C. A., Garlena, R. A., Daniel A. Russell, et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nature Medicine, 25(5), 730–733.
Schooley, R. T., Biswas, B., Gill, J. J., Hernandez-Morales, A., Lancaster, J., Lessor, L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrobial Agents and Chemotherapy, 61(10), e00954–e00917. https://doi.org/10.1007/s12010-020-03310-0.
Solovieva, E. V., Myakinina, V. P., Kislichkina, A. A., Krasilnikova, V. M., Verevkin, V. V., Mochalov, V. V., Lev, A. I., Fursova, N. K., & Volozhantsev, N. V. (2018). Comparative genome analysis of novel podoviruses lytic for hypermucoviscous Klebsiella pneumoniae of K1, K2, and K57 capsular types. Virus Research, 243, 10–18.
Tomat, D., Casabonnea, C., Aquilia, V., Balaguéa, C., & Quiberoni, A. (2018). Evaluation of a novel cocktail of six lytic bacteriophages against Shiga toxinproducing Escherichia coli in broth, milk and meat. Food Microbiology, 76, 434–442.
Kakasis, A., & Panitsa, G. (2019). Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Journal of Antimicrobial Agents, 53, 16–21.
Zhao, Y., Ye, M., Zhang, X., Sun, M., Zhang, Z., Chao, H., Huang, D., Wan, J., Zhang, S., Jiang, X., Sun, D., Yuan, Y., & Hu, F. (2019). Comparing polyvalent bacteriophage and bacteriophage cocktails for controlling antibiotic-resistant bacteria in soil-plant system. Science of the Total Environment, 657, 918–925.
El-Dougdoug, N. K., Cucic, S., Abdelhamid, A. G., Brovko, L., Kropinski, A. M., Griffiths, M. W., & Anany, H. (2019). Control of Salmonella Newport on cherry tomato using a cocktail of lytic bacteriophages. International Journal of Food Microbiology, 293, 60–71.
Soffer, N., Woolston, J., Li, M., et al. (2017). Bacteriophage preparation lytic for Shigella significantly reduces Shigella sonnei contamination in various foods. PLoS One, 12, e0175256.
Sváb, D., Falgenhauer, L., Rohde, M., Chakraborty, T., & Tóth, I. (2019). Complete genome sequence of C130_2, a novel myovirus infecting pathogenic escherichia coli and shigella strains. Archives of Virology, 164(1), 321–324.
Domingo-Calap, P., & Delgado-Martínez, J. (2018). Bacteriophages: Protagonists of a post-antibiotic era. Antibiotics., 7, 66.
Doore, S. M., Schrad, J., Dean, W. F., Dover, J. A., & Parent, K. N. (2018). Shigella phages isolated during a dysentery outbreak reveal uncommon structures and broad species diversity. Journal of Virology, 92, JVI.02117–JVI.02117.
Jurczak-Kurek, A., Gasior, T., Nejman-Falenczyk, B., Bloch, S., Dydecka, A., Topka, G., et al. (2016). Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Scientific Reports, 6, 34338.
Joseph, S., & David, W. R. (2001). Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Fan, N., Qi, R., & Yang, M. (2017). Isolation and characterization of a virulent bacteriophage infecting Acinetobacter johnsonii from activated sludge. Research in Microbiology, 5, 472–481.
Turner, D., Reynolds, D., Seto, D., & Mahadevan, P. (2013). Coregenes3.5: a webserver for the determination of core genes from sets of viral and small bacterial genomes. BMC Research Notes, 6(1), 140.
Pettengill, J. B., Luo, Y., Davis, S., Chen, Y., & Strain, E. (2014). An evaluation of alternative methods for constructing phylogenies from whole genome sequence data: A case study with salmonella. PeerJ., 2(6), e620.
Yoda, T., Tanabe, M., Tsuji, T., et al. (2017). Exonuclease processivity of archaeal replicative DNA polymerase in association with PCNA is expedited by mismatches in DNA. Scientific Reports-UK, 7, 44582.
Konomura, N., Arai, N., Shinohara, T., et al. (2017). Rad51 and RecA juxtapose dsDNA ends ready for DNA ligase-catalyzed end-joining under recombinase-suppressive conditions. Nucleic Acids Research, 45(1), 337–352.
Shlomai, J., & Linial, M. (1986). A nicking enzyme from trypanosomatids which specifically affects the topological linking of duplex DNA circles. Purification and characterization. The Journal of Biological Chemistry, 261(34), 16219–16225.
Jin, J., Li, Z. J., Wang, S. W., et al. (2014). Genome organisation of the Acinetobacter lytic phage ZZ1 and comparison with other T4-like Acinetobacter phages. BMC Genomics, 15(1), 793.
Young, R., & Blasi, U. (1995). Holins: Form and function in bacteriophage lysis. FEMS Microbiology Reviews, 17(1-2), 191–205.
Rui, H., Li, M., Tang, T., et al. (2018). Construction of Lactobacillus casei ghosts by Holin-mediated inactivation and the potential as a safe and effective vehicle for the delivery of DNA vaccines. BMC Microbiology, 18(1), 80.
Roach, D. R., & Donovan, D. M. (2015). Antimicrobial bacteriophage derived proteins and therapeutic applications. Bacteriophage., 5(3), e1062590.
Nikhat, Z., Raja, M., & Donald, S. (2002). CoreGenes: A computational tool for identifying and cataloging "core" genes in a set of small genomes. BMC Bioinformatics, 3(1), 12–12.
Acknowledgements
The authors would like to acknowledge the Institute of Microbiology, Chinese Academy of Sciences for electron microscopy.
Funding
This study was funded by grant from the National Natural Science Foundation of China (No. 50978250 and No. 51378485).
Author information
Authors and Affiliations
Contributions
H. Lu and H. Liu performed the experiments, analyzed the data, and wrote the paper. ML helped design the experiment and put forward suggestions for the smooth progress of the experiment. JW reviewed the first draft of the paper. XL and RL revised the manuscript. All authors had read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for Publication
Not applicable.
Conflict of Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 82 kb)
Rights and permissions
About this article
Cite this article
Lu, H., Liu, H., Lu, M. et al. Isolation and Characterization of a Novel myovirus Infecting Shigella dysenteriae from the Aeration Tank Water. Appl Biochem Biotechnol 192, 120–131 (2020). https://doi.org/10.1007/s12010-020-03310-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03310-0