Abstract
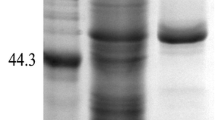
cDNA coding a prolyl aminopeptidase (PAP) was cloned from Aspergillus oryzae and over expressed in Bacillus subtilis with a 6×His tag in N-terminus. The recombinant prolyl aminopeptidase was secreted to extracellular by adding 2 mM CaCl2 and 5% D-sorbitol in TB medium; the enzyme activity in fermented supernatant increased from 7.2 to 41.5 U mL−1. It has been purified 4.3-fold through Ni-chelating affinity chromatography with a recovery of 47.3%. The purified enzyme is stable below 50 °C and within pH 6–11, and with the highest activity at pH 7.5 and 50 °C. Several kinds of salt can activate enzyme activity in a certain concentration and the relative activity was 127.02% even when the concentration of NaCl reached 4.36 M. It cleaved N-terminal Pro residues from many peptides but shown different hydrolysis rates for various Pro-X dipeptides or peptides which are of different lengths. It combined with alkaline protease and leucine aminopeptidase to hydrolyze casein, many free amino acid especially proline and small peptide of hydrolysate increased significantly.



Similar content being viewed by others
References
Gonzales, T., & Robert-Baudouy, J. (1996). Bacterial aminopeptidases: properties and functions. FEMS Microbiology Reviews, 18, 319–344.
Gao, X. X., Cui, W. J., Tian, Y. P., & Zhou, Z. M. (2013). Over-expression, secretion, biochemical characterisation, and structure analysis of Bacillus subtilis aminopeptidase. Journal of the Science of Food & Agriculture, 93, 2810–2815.
Santos, K., & Medrano, F. J. (2007). Expression, purification, and characterization of an aminopeptidase (Xac2987) with broad specificity from Xanthomonas axonopodis pv. Citri. Protein Expression and Purification, 52, 117–122.
Nicole, M., Holger, Z., & Martin, R. (2015). Prolyl-specifific peptidases for applications in food protein hydrolysis. Applied Microbiology and Biotechnology, 99, 7837–7846.
Nandan, A., Pandey, A., & Nampoothiri, K. M. (2011). Proline-specific extracellular aminopeptidase purified from Streptomyces lavendulae. Applied biochemistry & biotechnology, 163, 994–1001.
Polgar, L. (2005). The catalytic triad of serine peptidases. Cellular and Molecular Life Sciences, 62, 2167–2172.
Li, M., Chen, C. A., Davies, D. R., & Chin, T. K. (2010). Induced-fit mechanism for prolyl endopeptidase. Journal of Biological Chemistry, 285, 21487–21495.
Sarid, S., Berger, A., & Katchalski, E. (1959). Proline iminopeptidase. Journal of Biological Chemistry, 234, 1740–1746.
Matsushita-Morita, M., Furukawa, I., Suzuki, S., Yamagata, Y., Koide, Y., Ishida, H., Takeuchi, M., Kashiwagi, Y., & Kusumoto, K. I. (2010). Characterization of recombinant prolyl aminopeptidase from Aspergillus oryzae. Journal of Applied Microbiology, 109, 156–165.
Li, N., Wu, J. M., Zhang, L. F., Feng, H., et al. (2010). Characterization of a unique proline iminopeptidase from white-rot basidiomycetes Phanerochaete chrysosporium. Biochimie, 92, 779–788.
Arima, J., Uesugi, Y., Iwabuchi, M., & Hatanaka, T. (2008). Streptomyces aminopeptidase P: biochemical characterization and insight into the roles of its N-terminal domain. Protein Engineering,Design & Selection, 21, 45–53.
Murai, A., Tsujimoto, Y., Matsui, H., & Watanabe, K. (2004). An Aneurinibacillus sp. strain AM-1 produces a proline-specific aminopeptidase useful for collagen degradation. Journal of Applied Microbiology, 96, 810–818.
Akioka, M., Nakano, H., Horikiri, A., Tsujimoto, Y., Matsui, H., Shimizu, T., Nakatsu, T., Kato, H., & Watanabe, K. (2006). Overexpression, purification, crystallization and preliminary X-ray crystallographic studies of a proline-specific aminopeptidase from Aneurinibacillus sp. strain AM-1. Acta Crystallographica Section F-Structural Biology and Crystallization Communications, 62, 1266–1268.
Zhang, J., Kang, Z., Ling, Z., Cao, W., Liu, L., Wang, M., Du, G., & Chen, J. (2013). High-level extracellular production of alkaline polygalacturonate lyase in Bacillus subtilis with optimized regulatory elements. Bioresource Technology, 146, 543–548.
Harwood, C. R., & Cranenburgh, R. (2008). Bacillus protein secretion: an unfolding story. Trends in Microbiology, 16, 73–79.
Ploss, T. N., Reilman, E., Montefreeante, C. G., et al. (2016). Homogeneity and heterogeneity in amylase production by Bacillus subtilis under different growth conditions. Microbial Cell Factories, 15, 57.
Barbe, V., Cruveiller, S., Kunst, F., et al. (2009). From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology-SGM., 155, 1758–1775.
Kawabata, Y., Kimara, K., & Funane, K. (2012). Extracellular production of cycloisomaltooligosaccharide glucanotransferase and cyclodextran by a protease-deficient Bacillus subtilis host–vector system. Applied Microbiology and Biotechnology, 93, 1877–1884.
Vavrova, L., Muchova, K., & Barak, I. (2010). Comparison of different Bacillus subtilis expression systems. Reserach Microbiology., 146, 791–797.
Zweers, J. C., Barak, I., Becher, D., Driessen, A. J., Hecker, M., Kontinen, V. P., Saller, M. J., Vavrova, L., & Dijl, J. M. V. (2008). Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microbial Cell Factories, 7, 1–20.
Dijl, J. M. V., & Hecker, M. (2013). Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microbial Cell Factories, 12, 3–3.
Liu, B., Zhang, J., Li, B., Liao, X. R., Du, G. C., & Chen, J. (2013). Expression and characterization of extreme alkaline, oxidation-resistant keratinase from Bacillus licheniformis in recombinant Bacillus subtilis WB600 expression system and its application in wool fiber processing. World Journal of Microbiology & Biotechnology, 29, 825–832.
Dong, H. N., & Zhang, D. W. (2014). Current development in genetic engineering strategies of Bacillus species. Microbial Cell Factories, 13, 63.
Ding, G. W., Zhou, N. D., & Tian, Y. P. (2014). Over-expression of a proline specific aminopeptidase from Aspergillus oryzae JN-412 and its application in collagen degradation. Applied Biochemistry & Biotechnology, 173, 1765–1777.
Liu, J. M., Xin, X. J., Li, C. X., Xu, J. H., & Bao, J. (2012). Cloning of thermostable cellulase genes of Clostridium thermocellum and their secretive expression in Bacillus subtilis. Applied Biochemistry & Biotechnology, 166, 652–662.
Atlan, D., Laloi, P., & Portalier, R. (1989). Isolation and characterization of aminopeptidase-deficient Lactobacillus bulgaricus mutants. Applied and Environmental Microbiology, 55, 1717–1723.
Fuke, Y., & Matsuoka, H. (1993). The purification and characterization of prolyl aminopeptidase from Penicillium camemberti. Journal of Dairy Science, 76, 2478–2484.
Yu, Z., Wang, Q., Ma, Q., & Zhang, R. (2013). Secretory expression of lacticin Q fused with SUMO in Bacillus subtilis. Protein Expression & Purification, 89, 51–55.
Rawlings, N. D., Barrett, A. J., & Bateman, A. (2010). MEROPS: the peptidase database. Nucleic Acids Research, 36, 320–325.
Basten, D. E., Moers, A. P., Ooyen, A. J., & Schaap, P. J. (2005). Characterisation of Aspergillus niger prolyl aminopeptidase. Molecular Genetics & Genomics, 272, 673–679.
Mahon, C. S., Goetz, D. H., Tuohy, M. G., et al. (2009). Characterization of a multimeric, eukaryotic prolyl aminopeptidase: an inducible and highly specific intracellular peptidase from the non-pathogenic fungus Talaromyces emersonii. Microbiology-SGM, 155, 3673–3682.
Wang, Y., Liu, H., Wang, S., Li, H., & Xin, Q. (2015). Overexpressing of a novel wheat prolyl aminopeptidase gene enhances zinc stress tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue & Organ Culture, 121, 489–499.
Uraji, M., Arima, J., Uesugi, Y., Iwabuchi, M., & Hatanaka, T. (2007). Effect of salt on the activity of Streptomyces prolyl aminopeptidase. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, 1774, 1462–1469.
Fukuchi, S., Yoshimune, K., Wakayama, M., Moriguchi, M., & Nishikawa, K. (2003). Unique amino acid composition of proteins in halophilic bacteria. Journal of Molecular Biology, 327, 347–357.
Gao, X. X., Cui, W. J., Ding, N., Liu, Z. M., Tian, Y. P., & Zhou, Z. M. (2013). Structure-based approach to alter the substrate specificity of Bacillus subtilis aminopeptidase. Prion, 7, 328–334.
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 Program, 2011AA100905), the Fundamental Research Funds for the Central Universities (JUSRP51402A), and the Program for New Century Excellent Talents in University (NCET-12-0878).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(DOCX 624 kb)
Rights and permissions
About this article
Cite this article
Wang, KD., Wang, KH., Zhou, ND. et al. Secretory Expression, Purification, Characterization, and Application of an Aspergillus oryzae Prolyl Aminopeptidase in Bacillus subtilis . Appl Biochem Biotechnol 181, 1611–1623 (2017). https://doi.org/10.1007/s12010-016-2305-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2305-3