Abstract
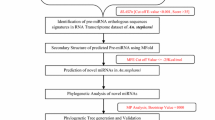
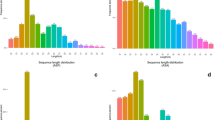
For studies on functional genomics, small RNAs, especially microRNAs (miRNAs), have emerged as a hot topic due to their importance in cellular and developmental processes. Identification of insect miRNAs largely depends on the availability of genomic sequences in the public domain. The large milkweed bug, Oncopeltus fasciatus (Dallas) is a hemimetabolous insect which has become a model hemipteran system for various molecular studies. In this study, we identified 96 candidate mature miRNAs from O. fasciatus genome using a blast search with the previously reported animal miRNAs. The secondary structure of predicted miRNA sequences was determined online using “mfold” web server and verified by calculating the minimal free energy index (MFEI). Six miRNAs let-7e, miR-133c, miR-219b, mir-466d, mir-669f, and mir-669l are reported for the first time in Insecta. Comparison of O. fasciatus mir-2 and mir-71 family clusters to those of diverse insect species showed that they are highly conserved. The phylogenetic analysis of miRNAs revealed the evolutionary relationship of conserved miRNAs of O. fasciatus with other insect species. Using a classical rule-based algorithm method, we predicted the possible targets of the new miRNAs. Our study not only identified the list of miRNAs in O. fasciatus but also provides a basic platform for developing novel pest management strategies based on artificial miRNAs.





Similar content being viewed by others
References
Asgari, S. (2013). MicroRNA functions in insects. Insect Biochemistry and Molecular Biology, 43, 388–397.
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., & Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858.
Agrawal, N., Sachdev, B., Rodrigues, J., Sree, K. S., & Bhatnagar, R. K. (2013). Development associated profiling of chitinase and microRNA of Helicoverpa armigera identified chitinase repressive microRNA. Scientific Reports, 3, 2292.
Zeng, Y., & Cullen, B. R. (2005). Efficient processing of primary microRNA hairpins by Drosha requires flanking non-structured RNA sequences. Journal of Biological Chemistry, 280, 27595–27603.
Winter, J., & Diederichs, S. (2011). Argonaute proteins regulate microRNA stability: increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biology, 8, 1149–1157.
Lee, I., Ajay, S. S., Yook, J. I., Kim, H. S., Hong, S. H., Kim, N. H., Dhanasekaran, S. M., Chinnaiyan, A. M., & Athey, B. D. (2009). New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Research, 19, 1175–1183.
Jagadeeswaran, G., Zheng, Y., Sumathipala, N., Jiang, H., Arrese, E. L., Soulages, J. L., Zhang, W., & Sunkar, R. (2010). Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics, 11, 52.
Alves e Silva, T. L., Vasconcellos, L. R. C., Lopes, A. H., & Souto-Padrón, T. (2013). The immune response of hemocytes of the insect Oncopeltus fasciatus against the flagellate Phytomonas serpens. PLoS ONE, 8, e72076.
Liu, J., Lemonds, T. R., & Popadic, A. (2014). The genetic control of aposematic black pigmentation in hemimetabolous insects: insights from Oncopeltus fasciatus. Evolution & Development, 16, 270–277.
Ewen-Campen, B., Shaner, N., Panfilio, K. A., Suzuki, Y., Roth, S., & Extavour, C. G. (2011). The maternal and early embryonic transcriptome of the large milkweed bug Oncopeltus fasciatus. BMC Genomics, 12, 61.
Qu, J., Richards, S., Bandaranaike, D., Bellair, M., Blankenburg, K., Chao, H., Dinh, H., Doddapaneni, H., Downs, B., Dugan-Rocha, S., Elkadiri, S., Gnanaolivu, R., Hernandez, B., Javaid, M., Jayaseelan, J. C., Lee, S., Li, M., Ming, W., Munidasa, M., Muniz, J., Nguyen, L., Ongeri, F., Osuji, N., Pu, L. L., Puazo, M., Qu, C., Quiroz, J., Raj, R., Weissenberger, G., Xin, Y., Zou, X., Han, Y., Worley, K., Muzny, D., & Gibbs, R. (2014). NCBI-BioProject: PRJNA229125, whole genome assembly of oncopeltus fasciatus using multiple sequencing technologies (http://www.ncbi.nlm.nih.gov/bioproject/229125).
Kozomara, A., & Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research, 42, D68–D73.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Ellango, R., Asokan, R., Mahmood, R., Ramamurthy, V. V., & Krishnakumar, N. K. (2014). Isolation of new microRNAs from the diamondback moth (Lepidoptera: yponomeutidae) genome by a computational method. Florida Entomologist, 97, 877–885.
Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31, 3406–3415.
Singh, J., & Nagaraju, J. (2008). Insilco prediction and characterization of microRNAs from red flour beetle (Tribolium castaneum). Insect Molecular Biology, 17, 427–436.
Ghosh, Z., Chakrabarti, J., & Mallick, B. (2007). miRNomics—the bioinformatics of microRNA genes. Biochemistry Biophysics Research Communication, 363, 6–11.
Zhang, B. H., Pan, X. P., Cannon, C. H., Cobb, G. P., & Anderson, T. A. (2006). Computational identification of microRNAs and their targets. Computational Biology and Chemistry, 30, 395–407.
Griffiths-Jones, S., Grocock, R. J., Van Dongen, S., Bateman, A., & Enright, A. J. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research, 34, D140–D144.
Griffiths-Jones, S. (2004). The microRNA registry. Nucleic Acids Research, 32, D109–D111.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C., & Marks, D. S. (2003). MicroRNA targets in Drosophila. Genome Biology, 5, R1.
Rehmsmeier, M., Steffen, P., Hoechsmann, M., & Giegerich, R. (2004). Fast and effective prediction of microRNA/target duplexes RNA. RNA, 10, 1507–1517.
Legeai, F., Rizk, G., Walsh, T., Edwards, O., Gordon, K., Lavenier, D., Leterme, N., Mereau, A., Nicolas, J., Tagu, D., & Jaubert-Possamai, S. (2010). Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genomics, 11, 281.
Lau, N. C., Lim, L. P., Weinstein, E. G., & Bartel, D. P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862.
Altuvia, Y., Landgraf, P., Lithwick, G., Elefant, N., Pfeffer, S., Aravin, A., Brownstein, M. J., Tuschl, T., & Margalit, H. (2005). Clustering and conservation patterns of human microRNAs. Nucleic Acids Research, 33, 2697–2706.
Saini, H. K., Enright, A. J., & Griffiths-Jones, S. (2008). Annotation of mammalian primary microRNAs. BMC Genomics, 9, 564.
Marco, A., Hui, J. H., Ronshaugen, M., & Griffiths-Jones, S. (2010). Functional shifts in insect microRNA evolution. Genome Biology and Evolution, 2, 686–696.
Biryukova, I., Ye, T., & Levashina, E. (2014). Transcriptome-wide analysis of microRNA expression in the malaria mosquito Anopheles gambiae. BMC Genomics, 15, 557.
Shin, C., Nam, J.-W., Farh, K. K.-H., Chiang, H. R., Shkumatava, A., & Bartel, D. P. (2010). Expanding the microRNA targeting code: functional sites with centered pairing. Molecular Cell, 38, 789–802.
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell, 23, 215–233.
Rigoutsos, I. (2009). New tricks for animal microRNAs: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Research, 69, 3245–3248.
Acknowledgments
Our sincere thanks are due to the Director, Indian Institute of Horticultural Research, Bengaluru, India, for providing the facilities and encouragement. We gratefully acknowledge the financial support received from the Indian Council of Agricultural Research (ICAR), New Delhi through the XIIth plan Network Project on ORP-on Management of sucking pest on horticultural crops.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Oncopeltus fasciatus mature miRNA details. (XLS 47 kb)
Table S2
O. fasciatus precursor miRNA details. (XLS 39 kb)
Table S3
Prediction of possible targets of O. fasciatus miRNA. (XLS 32 kb)
Table S4
Both Miranda and RNA hybrid were used to predict the possible target for six ofa-miRNAs. (XLS 75 kb)
Rights and permissions
About this article
Cite this article
Ellango, R., Asokan, R. & Ramamurthy, V.V. Insilco Prediction and Characterization of microRNAs from Oncopeltus fasciatus (Hemiptera: Lygaeidae) Genome. Appl Biochem Biotechnol 179, 1393–1403 (2016). https://doi.org/10.1007/s12010-016-2072-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2072-1