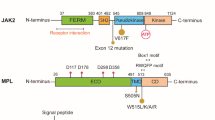
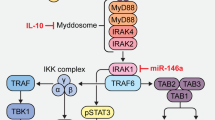
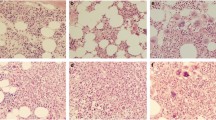
Abstract
Purpose of Review
Myeloproliferative neoplasms (MPNs) are chronic hematological malignancies characterized by increased proliferation of MPN stem and myeloid progenitor cells with or without bone marrow fibrosis that typically lead to increased peripheral blood cell counts. The genetic and cytogenetic alterations that initiate and drive the development of MPNs have largely been defined, and we summarize these here.
Recent Findings
In recent years, advances in understanding the pathogenesis of MPNs have defined a long-preclinical phase in JAK2-mutant MPN, identified genetic loci associated with MPN predisposition and uncovered mechanistic insights in CALR-mutant MPN. The integration of molecular genetics into prognostic risk models is well-established in myelofibrosis and ongoing studies are interrogating the prognostic implications of concomitant mutations in ET and PV. Despite all these advances, the field is deficient in clonally selective therapies to effectively target the MPN clone at any stage of disease, from pre-clinical to advanced.
Summary
Although the biological understanding of the pathogenesis of MPNs has progressed quickly, substantial knowledge gaps remain, including in the molecular mechanisms underlying MPN progression and myelofibrotic transformation. An ongoing goal for the MPN field is to translate advances in biological understanding to improved treatments for patients.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Dameshek W. Some speculations on the myeloproliferative syndromes editorial. Blood. 1951;6(4):372–375. Blood. 2016;127:663. https://doi.org/10.1182/blood-2015-12-686402.
Mullally A, Lane SW, Ball B, Megerdichian C, Okabe R, Al-Shahrour F, et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell. 2010;17:584–96. https://doi.org/10.1016/j.ccr.2010.05.015.
Li J, Prins D, Park HJ, Grinfeld J, Gonzalez-Arias C, Loughran S, et al. Mutant calreticulin knockin mice develop thrombocytosis and myelofibrosis without a stem cell self-renewal advantage. Blood. 2018;131:649–61. https://doi.org/10.1182/blood-2017-09-806356.
Grinfeld J, Nangalia J, Baxter EJ, Wedge DC, Angelopoulos N, Cantrill R, et al. Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 2018;379:1416–30. https://doi.org/10.1056/NEJMoa1716614. This study of more than 2000 patients with MPN integrated 63 clinical and genetic variables to identify distinct (genetic) subgroups that predict outcomes in MPN patients.
Thompson ER, Nguyen T, Kankanige Y, Yeh P, Ingbritsen M, McBean M, et al. Clonal independence of JAK2 and CALR or MPL mutations in comutated myeloproliferative neoplasms demonstrated by single cell DNA sequencing. Haematologica. 2021;106:313–5. https://doi.org/10.3324/haematol.2020.260448.
James C, Ugo V, Le Couédic J-P, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–8. https://doi.org/10.1038/nature03546.
Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–61. https://doi.org/10.1016/S0140-6736(05)71142-9.
Kralovics R. Acquired uniparental disomy of chromosome 9p is a frequent stem cell defect in polycythemia vera. Exp Hematol. 2002;30:229–36. https://doi.org/10.1016/S0301-472X(01)00789-5.
Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJP, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–97. https://doi.org/10.1016/j.ccr.2005.03.023.
Glassman CR, Tsutsumi N, Saxton RA, Lupardus PJ, Jude KM, Garcia KC. Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science. 2022;376:163–9. https://doi.org/10.1126/science.abn8933. This study employed cryo-electron microscopy to uncover a three-dimensional structure of the Janus kinase receptor.
Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–68. https://doi.org/10.1056/NEJMoa065202.
Hu X, li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Sig Transduct Target Ther. 2021;6:402. https://doi.org/10.1038/s41392-021-00791-1.
Wernig G, Gonneville JR, Crowley BJ, Rodrigues MS, Reddy MM, Hudon HE, et al. The Jak2V617F oncogene associated with myeloproliferative diseases requires a functional FERM domain for transformation and for expression of the Myc and Pim proto-oncogenes. Blood. 2008;111:3751–9. https://doi.org/10.1182/blood-2007-07-102186.
Meyer SC, Ghosh N, Stivala S, Baerenwaldt A, Hao-Shen H, Dirnhofer S, et al. Targeting cell non-autonomous MAPK activation as a novel therapeutic strategy in myeloproliferative neoplasms. Blood. 2017;130:381. https://doi.org/10.1182/blood.V130.Suppl_1.381.381.
Aranaz P, Hurtado C, Erquiaga I, Miguéliz I, Ormazábal C, Cristobal I, et al. CBL mutations in myeloproliferative neoplasms are also found in the gene’s proline-rich domain and in patients with the V617FJAK2. Haematologica. 2012;97:1234–41. https://doi.org/10.3324/haematol.2011.052605.
Fourouclas N, Li J, Gilby DC, Campbell PJ, Beer PA, Boyd EM, et al. Methylation of the suppressor of cytokine signaling 3 gene (SOCS3) in myeloproliferative disorders. Haematologica. 2008;93:1635–44. https://doi.org/10.3324/haematol.13043.
Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–405. https://doi.org/10.1056/NEJMoa1312542.
Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–90. https://doi.org/10.1056/NEJMoa1311347.
Fucikova J, Spisek R, Kroemer G, Galluzzi L. Calreticulin and cancer. Cell Res. 2021;31:5–16. https://doi.org/10.1038/s41422-020-0383-9.
How J, Hobbs GS, Mullally A. Mutant calreticulin in myeloproliferative neoplasms. Blood. 2019;134:2242–8. https://doi.org/10.1182/blood.2019000622.
Tefferi A, Lasho TL, Finke C, Belachew AA, Wassie EA, Ketterling RP, et al. Type 1 vs type 2 calreticulin mutations in primary myelofibrosis: differences in phenotype and prognostic impact. Leukemia. 2014;28:1568–70. https://doi.org/10.1038/leu.2014.83.
Pecquet C, Chachoua I, Roy A, Balligand T, Vertenoeil G, Leroy E, et al. Calreticulin mutants as oncogenic rogue chaperones for TpoR and traffic-defective pathogenic TpoR mutants. Blood. 2019;133:2669–81. https://doi.org/10.1182/blood-2018-09-874578.
Elf S, Abdelfattah NS, Chen E, Perales-Patón J, Rosen EA, Ko A, et al. Mutant calreticulin requires both its mutant C-terminus and the thrombopoietin receptor for oncogenic transformation. Cancer Discov. 2016;6:368–81. https://doi.org/10.1158/2159-8290.CD-15-1434.
Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M, et al. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood. 2006;108:3472–6. https://doi.org/10.1182/blood-2006-04-018879.
Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3:e270. https://doi.org/10.1371/journal.pmed.0030270.
Tefferi A, Lasho TL, Finke CM, Knudson RA, Ketterling R, Hanson CH, et al. CALR vs JAK2 vs MPL-mutated or triple-negative myelofibrosis: clinical, cytogenetic and molecular comparisons. Leukemia. 2014;28:1472–7. https://doi.org/10.1038/leu.2014.3.
Tefferi A, Nicolosi M, Mudireddy M, Szuber N, Finke CM, Lasho TL, et al. Driver mutations and prognosis in primary myelofibrosis: Mayo-Careggi MPN alliance study of 1,095 patients. Am J Hematol. 2018;93:348–55. https://doi.org/10.1002/ajh.24978.
Landgren O, Goldin LR, Kristinsson SY, Helgadottir EA, Samuelsson J, Björkholm M. Increased risks of polycythemia vera, essential thrombocythemia, and myelofibrosis among 24,577 first-degree relatives of 11,039 patients with myeloproliferative neoplasms in Sweden. Blood. 2008;112:2199–204. https://doi.org/10.1182/blood-2008-03-143602.
Maffioli M, Genoni A, Caramazza D, Mora B, Bussini A, Merli M, et al. Looking for CALR mutations in familial myeloproliferative neoplasms. Leukemia. 2014;28:1357–60. https://doi.org/10.1038/leu.2014.33.
Hinds DA, Barnholt KE, Mesa RA, Kiefer AK, Do CB, Eriksson N, et al. Germ line variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood. 2016;128:1121–8. https://doi.org/10.1182/blood-2015-06-652941.
Elbracht M, Meyer R, Kricheldorf K, Gezer D, Thomas E, Betz B, et al. Germline variants in DNA repair genes, including BRCA1/2, may cause familial myeloproliferative neoplasms. Blood Adv. 2021;5:3373–6. https://doi.org/10.1182/bloodadvances.2021004811.
Jones AV, Chase A, Silver RT, Oscier D, Zoi K, Wang YL, et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat Genet. 2009;41:446–9. https://doi.org/10.1038/ng.334.
Kilpivaara O, Mukherjee S, Schram AM, Wadleigh M, Mullally A, Ebert BL, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41:455–9. https://doi.org/10.1038/ng.342.
Olcaydu D, Harutyunyan A, Jäger R, Berg T, Gisslinger B, Pabinger I, et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet. 2009;41:450–4. https://doi.org/10.1038/ng.341.
Bao EL, Nandakumar SK, Liao X, Bick AG, Karjalainen J, Tabaka M, et al. Inherited myeloproliferative neoplasm risk affects haematopoietic stem cells. Nature. 2020;586:769–75. https://doi.org/10.1038/s41586-020-2786-7. This large-scale genome-wide association study identified 17 genetic loci associated with inherited risk of MPN.
Titmarsh GJ, Duncombe AS, McMullin MF, O’Rorke M, Mesa R, de Vocht F, et al. How common are myeloproliferative neoplasms? A systematic review and meta-analysis. Am J Hematol. 2014;89:581–7. https://doi.org/10.1002/ajh.23690.
Cordua S, Kjaer L, Skov V, Pallisgaard N, Hasselbalch HC, Ellervik C. Prevalence and phenotypes of JAK2 V617F and calreticulin mutations in a Danish general population. Blood. 2019;134:469–79. https://doi.org/10.1182/blood.2019001113. This study used droplet digital PCR to screen 19,958 individuals from the Danish population and identified a prevalence of 3.5% for the JAK2V617F mutation and 0.16% for CALR mutations.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98. https://doi.org/10.1056/NEJMoa1408617.
Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. https://doi.org/10.1056/NEJMoa1409405.
Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–21. https://doi.org/10.1056/NEJMoa1701719.
Wolach O, Sellar RS, Martinod K, Cherpokova D, McConkey M, Chappell RJ, et al. Increased neutrophil extracellular trap formation promotes thrombosis in myeloproliferative neoplasms. Sci Transl Med. 2018. https://doi.org/10.1126/scitranslmed.aan8292.
Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367:1449–54. https://doi.org/10.1126/science.aay9333.
Williams N, Lee J, Mitchell E, Moore L, Baxter EJ, Hewinson J, et al. Life histories of myeloproliferative neoplasms inferred from phylogenies. Nature. 2022;602:162–8. https://doi.org/10.1038/s41586-021-04312-6. This study used whole-genome sequencing to determine clonal histories in MPN patients and found that the mean latency between acquisition of a JAK2V617F mutation and development of MPN was 30 years.
van Egeren D, Escabi J, Nguyen M, Liu S, Reilly CR, Patel S, et al. Reconstructing the lineage histories and differentiation trajectories of individual cancer cells in myeloproliferative neoplasms. Cell Stem Cell. 2021;28:514–523.e9. https://doi.org/10.1016/j.stem.2021.02.001. This study found that the JAK2V617F mutation occurs in a single HSC several decades before MPN diagnosis and that. JAK2V617F-mutant HSCs have an increased fitness advantage.
Tefferi A, Guglielmelli P, Larson DR, Finke C, Wassie EA, Pieri L, et al. Long-term survival and blast transformation in molecularly annotated essential thrombocythemia, polycythemia vera, and myelofibrosis. Blood. 2014;124:2507–13; quiz 2615. :https://doi.org/10.1182/blood-2014-05-579136.
Tefferi A, Lasho TL, Tischer A, Wassie EA, Finke CM, Belachew AA, et al. The prognostic advantage of calreticulin mutations in myelofibrosis might be confined to type 1 or type 1-like CALR variants. Blood. 2014;124:2465–6. https://doi.org/10.1182/blood-2014-07-588426.
Cerquozzi S, Tefferi A. Blast transformation and fibrotic progression in polycythemia vera and essential thrombocythemia: a literature review of incidence and risk factors. Blood Cancer J. 2015;5:e366. https://doi.org/10.1038/bcj.2015.95.
Iurlo A, Cattaneo D, Gianelli U. Blast transformation in myeloproliferative neoplasms: risk factors, biological findings, and targeted therapeutic options. Int J Mol Sci. 2019. https://doi.org/10.3390/ijms20081839.
Guglielmelli P, Loscocco GG, Mannarelli C, Rossi E, Mannelli F, Ramundo F, et al. JAK2V617F variant allele frequency 50% identifies patients with polycythemia vera at high risk for venous thrombosis. Blood Cancer J. 2021;11:199. https://doi.org/10.1038/s41408-021-00581-6. This paper investigated the impact of JAK2V617F variant allele frequency (VAF) on MPN disease complications.
Rumi E, Pietra D, Ferretti V, Klampfl T, Harutyunyan AS, Milosevic JD, et al. JAK2 or CALR mutation status defines subtypes of essential thrombocythemia with substantially different clinical course and outcomes. Blood. 2014;123:1544–51. https://doi.org/10.1182/blood-2013-11-539098.
Akada H, Yan D, Zou H, Fiering S, Hutchison RE, Mohi MG. Conditional expression of heterozygous or homozygous Jak2V617F from its endogenous promoter induces a polycythemia vera-like disease. Blood. 2010;115:3589–97. https://doi.org/10.1182/blood-2009-04-215848.
Wang L, Swierczek SI, Lanikova L, Kim SJ, Hickman K, Walker K, et al. The relationship of JAK2(V617F) and acquired UPD at chromosome 9p in polycythemia vera. Leukemia. 2014;28:938–41. https://doi.org/10.1038/leu.2014.20.
Karantanos T, Chaturvedi S, Braunstein EM, Spivak J, Resar L, Karanika S, et al. Sex determines the presentation and outcomes in MPN and is related to sex-specific differences in the mutational burden. Blood Adv. 2020;4:2567–76. https://doi.org/10.1182/bloodadvances.2019001407.
Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, et al. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372:601–12. https://doi.org/10.1056/NEJMoa1412098.
Nangalia J, Nice FL, Wedge DC, Godfrey AL, Grinfeld J, Thakker C, et al. DNMT3A mutations occur early or late in patients with myeloproliferative neoplasms and mutation order influences phenotype. Haematologica. 2015;100:e438–42. https://doi.org/10.3324/haematol.2015.129510.
Tong J, Sun T, Ma S, Zhao Y, Ju M, Gao Y, et al. Hematopoietic stem cell heterogeneity is linked to the initiation and therapeutic response of myeloproliferative neoplasms. Cell Stem Cell. 2021;28:502–513.e6. https://doi.org/10.1016/j.stem.2021.01.018. This paper investigated the impact of HSC heterogeneity on MPN phenotype in the context of JAK2V617F-driven MPN.
Chen E, Beer PA, Godfrey AL, Ortmann CA, Li J, Costa-Pereira AP, et al. Distinct clinical phenotypes associated with JAK2V617F reflect differential STAT1 signaling. Cancer Cell. 2010;18:524–35. https://doi.org/10.1016/j.ccr.2010.10.013.
Duek A, Lundberg P, Shimizu T, Grisouard J, Karow A, Kubovcakova L, et al. Loss of Stat1 decreases megakaryopoiesis and favors erythropoiesis in a JAK2-V617F-driven mouse model of MPNs. Blood. 2014;123:3943–50. https://doi.org/10.1182/blood-2013-07-514208.
Grisouard J, Shimizu T, Duek A, Kubovcakova L, Hao-Shen H, Dirnhofer S, Skoda RC. Deletion of Stat3 in hematopoietic cells enhances thrombocytosis and shortens survival in a JAK2-V617F mouse model of MPN. Blood. 2015;125:2131–40. https://doi.org/10.1182/blood-2014-08-594572.
Marneth AE, Mullally A. The molecular genetics of myeloproliferative neoplasms. Cold Spring Harb Perspect Med. 2020. https://doi.org/10.1101/cshperspect.a034876.
Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. https://doi.org/10.1038/nature12213.
Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–8. https://doi.org/10.1182/blood-2013-11-537167.
Song J, Hussaini M, Zhang H, Shao H, Qin D, Zhang X, et al. Comparison of the mutational profiles of primary myelofibrosis, polycythemia vera, and essential thrombocytosis. Am J Clin Pathol. 2017;147:444–52. https://doi.org/10.1093/ajcp/aqw222.
Dunbar AJ, Rampal RK, Levine R. Leukemia secondary to myeloproliferative neoplasms. Blood. 2020;136:61–70. https://doi.org/10.1182/blood.2019000943.
Rahmani NE, Ramachandra N, Sahu S, Gitego N, Lopez A, Pradhan K, et al. ASXL1 mutations are associated with distinct epigenomic alterations that lead to sensitivity to venetoclax and azacytidine. Blood Cancer J. 2021;11:157. https://doi.org/10.1038/s41408-021-00541-0.
Yang H, Kurtenbach S, Guo Y, Lohse I, Durante MA, Li J, et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood. 2018;131:328–41. https://doi.org/10.1182/blood-2017-06-789669.
Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–93. https://doi.org/10.1016/j.ccr.2012.06.032.
Inoue D, Fujino T, Sheridan P, Zhang Y-Z, Nagase R, Horikawa S, et al. A novel ASXL1-OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia. 2018;32:1327–37. https://doi.org/10.1038/s41375-018-0083-3.
Newberry KJ, Patel K, Masarova L, Luthra R, Manshouri T, Jabbour E, et al. Clonal evolution and outcomes in myelofibrosis after ruxolitinib discontinuation. Blood. 2017;130:1125–31. https://doi.org/10.1182/blood-2017-05-783225.
Vannucchi AM, Lasho TL, Guglielmelli P, Biamonte F, Pardanani A, Pereira A, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27:1861–9. https://doi.org/10.1038/leu.2013.119.
Kröger N, Panagiota V, Badbaran A, Zabelina T, Triviai I, Araujo Cruz MM, et al. Impact of molecular genetics on outcome in myelofibrosis patients after allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2017;23:1095–101. https://doi.org/10.1016/j.bbmt.2017.03.034.
Luque Paz D, Riou J, Verger E, Cassinat B, Chauveau A, Ianotto J-C, et al. Genomic analysis of primary and secondary myelofibrosis redefines the prognostic impact of ASXL1 mutations: a FIM study. Blood Adv. 2021;5:1442–51. https://doi.org/10.1182/bloodadvances.2020003444.
Liang Y, Tebaldi T, Rejeski K, Joshi P, Stefani G, Taylor A, et al. SRSF2 mutations drive oncogenesis by activating a global program of aberrant alternative splicing in hematopoietic cells. Leukemia. 2018;32:2659–71. https://doi.org/10.1038/s41375-018-0152-7.
Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC-W, Ramakrishnan A, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–30. https://doi.org/10.1016/j.ccell.2015.04.006.
Smeets MF, Tan SY, Xu JJ, Anande G, Unnikrishnan A, Chalk AM, et al. Srsf2P95H initiates myeloid bias and myelodysplastic/myeloproliferative syndrome from hemopoietic stem cells. Blood. 2018;132:608–21. https://doi.org/10.1182/blood-2018-04-845602.
Wang Y-H, Lin C-C, Lee S-H, Tsai C-H, Wu S-J, Hou H-A, et al. ASXL1 mutation confers poor prognosis in primary myelofibrosis patients with low JAK2V617F allele burden but not in those with high allele burden. Blood Cancer J. 2020;10:99. https://doi.org/10.1038/s41408-020-00364-5.
Guglielmelli P, Gangat N, Coltro G, Lasho TL, Loscocco GG, Finke CM, et al. Mutations and thrombosis in essential thrombocythemia. Blood Cancer J. 2021;11:77. https://doi.org/10.1038/s41408-021-00470-y. This paper investigated the impact of mutational status on the risk of thrombosis in patients with Essential Thrombocythemia.
Tefferi A, Lasho TL, Guglielmelli P, Finke CM, Rotunno G, Elala Y, et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv. 2016;1:21–30. https://doi.org/10.1182/bloodadvances.2016000216.
Zhang S-J, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–5. https://doi.org/10.1182/blood-2011-11-390252.
Najfeld V, Montella L, Scalise A, Fruchtman S. Exploring polycythaemia vera with fluorescence in situ hybridization: additional cryptic 9p is the most frequent abnormality detected. Br J Haematol. 2002;119:558–66. https://doi.org/10.1046/j.1365-2141.2002.03763.x.
Gangat N, Strand J, Lasho TL, Finke CM, Knudson RA, Pardanani A, et al. Cytogenetic studies at diagnosis in polycythemia vera: clinical and JAK2V617F allele burden correlates. Eur J Haematol. 2008;80:197–200. https://doi.org/10.1111/j.1600-0609.2007.01003.x.
Wolanskyj AP, Gangat N, Schwager SM, Ketterling RP, Tefferi A. Cytogenetic abnormalities in essential thrombocythemia: prevalence and prognostic significance. Blood. 2006;108:3626. https://doi.org/10.1182/blood.V108.11.3626.3626.
Gangat N, Tefferi A, Thanarajasingam G, Patnaik M, Schwager S, Ketterling R, Wolanskyj AP. Cytogenetic abnormalities in essential thrombocythemia: prevalence and prognostic significance. Eur J Haematol. 2009;83:17–21. https://doi.org/10.1111/j.1600-0609.2009.01246.x.
Reilly JT, Snowden JA, Spearing RL, Fitzgerald PM, Jones N, Watmore A, Potter A. Cytogenetic abnormalities and their prognostic significance in idiopathic myelofibrosis: a study of 106 cases. Br J Haematol. 1997;98:96–102. https://doi.org/10.1046/j.1365-2141.1997.1722990.x.
Klampfl T, Harutyunyan A, Berg T, Gisslinger B, Passamonti F, Rumi E, et al. Chromosomal aberration network in myeloproliferative neoplasms. Blood. 2010;116:318. https://doi.org/10.1182/blood.V116.21.318.318.
Kralovics R, Passamonti F, Liu K, Teo S-S, Bench A, Tichelli A, et al. Loss of heterozygosity on chromosome 9p24 Is the most frequent chromosomal aberration in polycythemia vera and idiopathic myelofibrosis. Blood. 2004;104:2425. https://doi.org/10.1182/blood.V104.11.2425.2425.
Tefferi A, Nicolosi M, Mudireddy M, Lasho TL, Gangat N, Begna KH, et al. Revised cytogenetic risk stratification in primary myelofibrosis: analysis based on 1002 informative patients. Leukemia. 2018;32:1189–99. https://doi.org/10.1038/s41375-018-0018-z.
Gangat N, Caramazza D, Vaidya R, George G, Begna K, Schwager S, et al. DIPSS plus: a refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J Clin Oncol. 2011;29:392–7. https://doi.org/10.1200/JCO.2010.32.2446.
Guglielmelli P, Lasho TL, Rotunno G, Mudireddy M, Mannarelli C, Nicolosi M, et al. MIPSS70: mutation-enhanced international prognostic score system for transplantation-age patients with primary myelofibrosis. J Clin Oncol. 2018;36:310–8. https://doi.org/10.1200/JCO.2017.76.4886.
Hussein K, Pardanani AD, van Dyke DL, Hanson CA, Tefferi A. International Prognostic Scoring System-independent cytogenetic risk categorization in primary myelofibrosis. Blood. 2010;115:496–9. https://doi.org/10.1182/blood-2009-08-240135.
Strasser-Weippl K, Steurer M, Kees M, Augustin F, Tzankov A, Dirnhofer S, et al. Chromosome 7 deletions are associated with unfavorable prognosis in myelofibrosis with myeloid metaplasia. Blood. 2005;105:4146. https://doi.org/10.1182/blood-2004-11-4319.
Marcellino BK, Hoffman R, Tripodi J, Lu M, Kosiorek H, Mascarenhas J, et al. Advanced forms of MPNs are accompanied by chromosomal abnormalities that lead to dysregulation of TP53. Blood Adv. 2018;2:3581–9. https://doi.org/10.1182/bloodadvances.2018024018.
Supper E, Rudat S, Iyer V, Droop A, Wong K, Spinella J-F, et al. Cut-like homeobox 1 (CUX1) tumor suppressor gene haploinsufficiency induces apoptosis evasion to sustain myeloid leukemia. Nat Commun. 2021;12:2482. https://doi.org/10.1038/s41467-021-22750-8.
Acknowledgements
A.M. receives funding from the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH) (R01HL131835), the Gabrielle’s Angel Foundation, the Leukemia & Lymphoma Society and the Starr Cancer Consortium (grant #I15-0026). B.R. receives funding from the German Cancer Aid (Mildred-Scheel scholarship, grant #70114570).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
A.M. receives research funding from the Relay Therapeutics. A.M. has consulted for the Janssen, PharmaEssentia, Constellation, Aclaris Therapeutics, Cellarity, Morphic Therapeutics and Biomarin Pharmaceuticals. AM has received research funding from the Janssen and Actuate Therapeutics.
Human and Animal Rights and Informed Consent.
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Myeloproliferative Neoplasms
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rolles, B., Mullally, A. Molecular Pathogenesis of Myeloproliferative Neoplasms. Curr Hematol Malig Rep 17, 319–329 (2022). https://doi.org/10.1007/s11899-022-00685-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11899-022-00685-1