Abstract
Insulin resistance and the metabolic syndrome are complex metabolic traits and key risk factors for the development of cardiovascular disease. They result from the interplay of environmental and genetic factors but the full extent of the genetic background to these conditions remains incomplete. Large-scale genome-wide association studies have helped advance the identification of common genetic variation associated with insulin resistance and the metabolic syndrome, and more recently, exome sequencing has allowed the identification of rare variants associated with the pathogenesis of these conditions. Many variants associated with insulin resistance are directly involved in glucose metabolism; however, functional studies are required to assess the contribution of other variants to the development of insulin resistance. Many genetic variants involved in the pathogenesis of the metabolic syndrome are associated with lipid metabolism.
Similar content being viewed by others
Introduction
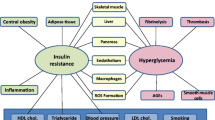
Both insulin resistance and the metabolic syndrome are powerful risk factors for the development of cardiovascular disease and type 2 diabetes (T2D). Insulin resistance can be defined as the inability of insulin to stimulate glucose disposal, and when an insulin-resistant individual is unable to secrete sufficient insulin to overcome this defect, T2D develops. The metabolic syndrome comprises a number of risk factors which occur together more often than by chance alone. A joint scientific statement issued in 2009 by the World Health Organisation (WHO), International Diabetes Federation (IDF), American Heart Association (AHA) and National Heart, Lung and Blood Institute (NHLBI) identified the metabolic syndrome as comprising hypertension, dyslipidaemia (raised triglyceride and low high-density lipoprotein cholesterol levels), hyperglycaemia and central obesity [1].
The prevalence of insulin resistance and the metabolic syndrome is increasing, particularly in developing countries and in younger populations with estimates of prevalence ranging from 20 to 40 % in different populations [2–4]. While there are strong lifestyle determinants for the development of both insulin resistance and the metabolic syndrome, it is increasingly clear that an individual’s risk of developing insulin resistance and aspects of the metabolic syndrome are also determined by genetic factors. Early familial genetic studies provided strong evidence for a genetic basis for both insulin resistance and the individual components of the metabolic syndrome [5–11], and since then, genome-wide association studies (GWAS) and, more recently, next-generation sequencing, have allowed for the identification of both common (defined by a minor allele frequency (MAF) >5 %) and rare (MAF < 0.5 %) genetic variants linked with these disease-associated traits. Associations of common genetic variation with a particular trait are generally defined with a genome-wide significance level of p < 5 × 10−8. The scope of this review is to summarise some of the genome-wide association studies (GWAS) studies and meta-analyses performed in relation to insulin resistance and the metabolic syndrome.
Common Genetic Variation in Insulin Resistance
Since 2007, genome-wide association studies (GWAS) have identified approximately 88 loci associated with risk of developing T2D [12]. The vast majority of the loci related to T2D are primarily associated with insulin secretion and β-cell function with far fewer variants apparently influencing insulin resistance. There are a number of reasons for this, not the least that insulin resistance is strongly linked to obesity, and dissecting out the role of common genetic variation in insulin resistance in the absence of obesity has been problematic. There has also been a lack of large cohorts with reliable measures of insulin sensitivity. Indeed, many measures examine whole body insulin sensitivity and do not measure insulin sensitivity at a tissue/organ level. Table 1 lists the loci and nearby genes associated with insulin resistance identified by linkage analysis, candidate gene studies and GWAS.
The GENESIS consortium is the only group to date that has published a GWAS of whole-body insulin sensitivity measured during hyperinsulinaemia (clamp/insulin suppression test) in a modest number of subjects (2764 with replication in an additional 2860 individuals) from four cohorts. The consortium found that variation in NAT2 was associated with insulin resistance although this did not reach formal genome-wide significance (rs1208 was associated with decreased insulin sensitivity with p = 9.81 × 10−7 after adjusting for BMI). The rs1208 loci was also associated with fasting glucose, triglycerides and total cholesterol and with an increase in coronary heart disease risk. A functional relationship between NAT2 and insulin sensitivity was confirmed in mouse 3T3-L1 adipocytes where NAT1 knockdown (the mouse orthologue of human NAT2) decreased insulin-stimulated glucose uptake and in a mouse NAT1 knockout model which showed decreased insulin sensitivity [13]. NAT2 is involved in acetylation and influences sensitivity to certain drugs such as isoniazid, although the endogenous substrates remain to be defined (reviewed in [14]). GWAS such as this are challenging to perform in large cohorts as the protocols involved in measuring whole-body insulin sensitivity are time-consuming, labour-intensive and, as such, expensive to perform.
The hyperinsulinaemic-euglycaemic clamp is considered the gold standard method for assessing insulin sensitivity (defined for these purposes as the ability of insulin to stimulate glucose uptake) but it is a long protocol and complex to perform. Simpler measures of insulin sensitivity such as fasting insulin concentrations and leptin/adiponectin ratios [15] are often used as a surrogate marker for insulin sensitivity. Fasting insulin and glucose levels can also be used to calculate the homeostasis model assessment (HOMA). HOMA-IR is used as a measure of insulin resistance which has the advantage of being simple to obtain but is not directly comparable to the clamp measure [16].
In an effort to identify new variants associated with insulin resistance, large-scale meta-analyses of GWAS have been performed using multiple cohorts which have measures of insulin sensitivity, processing and secretion. This approach has confirmed associations of many loci with specific glycaemic traits as well as identifying new loci. The Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) performed meta-analyses on GWAS from 21 cohorts of non-diabetic individuals including 46,186 individuals with measures of fasting glucose and 38,238 with measures of fasting insulin and HOMA-IR as an index of insulin resistance. Twenty-five SNPs were followed up in a further 76,558 individuals with this approach identifying 16 loci associated with fasting glucose and two with fasting insulin. This study confirmed loci near GCKR and a newly identified loci near IGF1 as being associated with insulin resistance [17] with these findings being replicated in a further 14 cohorts comprising 29,084 non-diabetic individuals with detailed measures of fasting proinsulin, insulin secretion and sensitivity [18]. This latter study has recently been expanded by Dimas and colleagues who examined the effect of 37 T2D risk loci on measures of insulin processing, secretion, sensitivity and clearance from up to 58,614 non-diabetic individuals with basal measures and 17,327 individuals with dynamic measures of glycaemic traits. They found that the risk loci could be subdivided into five clusters including one cluster with four loci, PPARγ, KLF14, IRS1 and GCKR, associated with insulin resistance [19].
More recently, alternate approaches to identifying variants associated with insulin resistance have been developed. Manning and colleagues have developed a joint meta-analysis (JMA) approach to identify SNPs significantly associated with either fasting glucose and/or fasting insulin while both adjusting for BMI and allowing for interaction with BMI [20]. This analysis method can increase the power to detect genetic associations where interaction with the environment is unknown [21, 22]. This approach identified six loci, including five new variants, as being associated with fasting insulin levels (IRS1, COBLL1-GRB14, PPP1R3B, PDGFC, UHRF1BP1 and LYPLAL1). Scott and colleagues used the Metabochip, a custom genotyping array containing 196,725 SNPs chosen due to their associations with a variety of cardiometabolic traits including type 2 diabetes, BMI and fasting glucose [23] and large-scale meta-analyses of up to 133,010 individuals to identify 17 loci significantly associated with fasting insulin. These loci included those previously identified by Manning, genes which have previously been associated with other metabolic traits (TCF7L2, PPARG, FTO, RSPO3, ANKRD55-MAP3K1 and ARL15) and newly identified loci (HIP1, TET2, YSK4, PEPD and FAM13A) [24]. These loci have been used in two further studies to generate an insulin resistance genetic risk score to examine the relationship between variants associated with fasting insulin and an individual’s risk of developing insulin resistance and T2D [25•, 26•]. Scott and colleagues generated an insulin resistance risk score from ten variants which were also specifically associated with lower HDL and higher triglycerides (IRS1, GRB14, ARL15, PPARG, PEPD, ANKRD55-MAP3K1, PDGFC, LYPLAL1, RSPO3 and FAM13A1) while Yaghootkar and colleagues selected 19 variants to generate their insulin resistance risk score. These studies found that the insulin resistance genetic risk score was associated with decreased insulin sensitivity and lower BMI [25•]. Yaghootkar and colleagues found that a cluster of 11 risk variants were associated with increased triglycerides and lower HDL-cholesterol along with a lower BMI, while individuals with 17 or more of these risk variants were slimmer but were at higher risk of developing T2D, coronary artery disease and hypertension compared with those individuals carrying nine or less of these insulin resistance risk variants [26•]. Together, these studies highlight that insulin resistance is not always associated with obesity and can occur in the absence of a high BMI.
Genetic Loci Associated with Insulin Resistance
Many of the loci that have been identified as being associated with risk of developing insulin resistance harbour genes nearby that are good biological candidates for association with measures of insulin sensitivity. The PPARγ (peroxisome proliferator-activated receptor gamma) variant Pro12Ala was one of the first genetic variants to be identified as being reproducibly associated with a decreased risk of developing T2D [27, 28]. PPARγ is a nuclear receptor involved in the transcription of genes involved in fatty acid and energy metabolism, and PPARy agonists are currently used in T2D treatment. rs972283 near KLF14 (kruppel-like factor 14) was identified through large-scale association analysis of a European cohort [29] associated with reduced insulin action, and KLF14 gene and protein expression have recently been shown to be significantly reduced in both adipose tissue and muscle from T2D patients [30]. IRS1 (insulin receptor substrate 1) is a key component of the insulin signalling pathway initiating the activation of PI3K in response to insulin. The C allele at rs2943641 adjacent to IRS1 was identified as being associated with insulin resistance and hyperinsulinaemia in a European population. Functional studies showed that the risk allele was associated with both lower basal IRS1 protein levels and reduced PI3K-activity during insulin infusion indicating a causative role for the variant on disease risk [31]. The SNP, rs2943650, near IRS1 has also been associated with a lower body fat percentage, increased triglycerides and insulin resistance, and lower HDL-cholesterol [32]. GCKR (glucokinase regulator) encodes glucokinase regulatory protein (GKRP) which binds to, and inhibits the activity of glucokinase, a key enzyme in regulating hepatic glucose disposal and storage [33].
Variants within the first intron of FTO were the first to be robustly linked with body mass index [34, 35]. Further studies found significant associations of the variants with obesity-related traits and also with fasting insulin and insulin sensitivity [36]. Variants within TCF7L2 (transcription factor 7-like 2) show the strongest and most consistent association with risk of developing T2D of any gene variants identified so far. The risk variant has been associated with both impaired beta cell function and insulin resistance [37–39]. Subsequent functional studies have focussed on the role of TCF7L2 in beta cell function [40, 41]. However, there is now increasing evidence for a role for TCF7L2 in insulin-dependent tissues [42]. A variant in NAT2 (N-acetyltransferase 2) was recently found to be the top signal in four European cohorts of non-diabetic individuals who underwent genome-wide genotyping as well as a direct measure of insulin sensitivity [13]. Functional analysis of the mouse orthologue, NAT1, demonstrated that NAT1 knockout mice had impaired insulin sensitivity in vivo.
Two loci near TMEM163 (transmembrane protein 163) were found to be associated with both reduced plasma insulin and HOMA-IR in a GWAS of a cohort with Indian ancestry [43]. IGF1 (insulin-like growth factor 1) is similar to insulin in function and regulates growth and development. Low plasma levels of IGF-1 have previously been associated with a reduction in insulin sensitivity [44], and analysis of the rs35767 polymorphism indicates that carriers of the G allele have lower circulating levels of IGF-1 compared to A allele carriers [45]. MC4R (melanocortin 4 receptor) was identified by GWAS of a UK cohort of Indian-Asian and European ancestry as being associated with both insulin resistance, as measured by HOMA-IR and waist circumference, with higher risk-allele frequencies being found in the Indian-Asian cohort [46]. Variants within MC4R have also been associated with BMI [47]. GWAS of an African-American cohort identified the SNP rs7077836 near TCERG1L (transcription elongation regulator 1-like) and rs17046216 in SC4MOL (sterol-C4-methyl oxidase-like) as being associated with both fasting insulin and HOMA-IR, a finding also replicated in a West African cohort [48]. ARL15 (ADP ribosylation factor like GTPase 15) belongs to a family of intracellular vesicle trafficking although its exact function is still unknown. Variants within ARL15 are associated with decreased adiponectin levels and nominally associated with risk of T2D, coronary heart disease and insulin resistance as measured by HOMA-IR [49]. Adiponectin is an adipokine involved in improving insulin sensitivity (reviewed in [50]). A recent study using a novel adiponectin receptor agonist suggests that activation of the adiponectin pathway directly improves insulin sensitivity [51].
Some of the novel loci are of unknown function but others, such as PPP1R3B (protein phosphatase 1 regulatory subunit 3B), are thought to play a role in skeletal muscle glycogen synthesis [52] while GRB14 (growth factor receptor bound protein 14) interacts with receptor tyrosine kinases such as the insulin and insulin-like growth factor receptors [53].
Common Genetic Variation in Metabolic Syndrome
The search for genetic determinants of the metabolic syndrome has largely taken two approaches, either examining the metabolic syndrome as a whole or as pairs of traits, or analysing associations with the individual components of the metabolic syndrome. Before GWAS, candidate gene association studies and linkage studies reported a number of potential genes associated with aspects of the metabolic syndrome but many of these findings were not replicated. Most GWAS to date have examined associations of variants with the individual components of the metabolic syndrome with at least 56 loci being reproducibly associated with obesity, 157 with lipids and over 90 loci associated with hypertension as well as the numerous loci associated with T2D (reviewed in [54]). It is beyond the scope of this review to discuss GWAS associated with the individual metabolic syndrome components. Instead, we focus on summarizing GWAS involving the examination of the metabolic syndrome as an entity in itself. Table 2 summarizes the variants identified in such studies.
The first GWAS examining common genetic variation in metabolic syndrome was performed on a population of Indian-Asian men who have a higher prevalence of metabolic syndrome compared to the European population. A total of approximately 317,000 SNPs were genotyped in 2700 individuals in stage 1 with the top 1500 SNPs followed up in 2300 individuals in stage 2. No common genetic basis for metabolic syndrome was identified in this study [55]. GWAS of a European population examined the association of approximately 2.5 million SNPs with either metabolic syndrome as a whole, or with pairs of traits in 22,161 individuals. Variants near BUD13, ZNF259, APOA5, LPL and CETP were found to be associated with metabolic syndrome, and a further 27 variants were associated with combinations of pairs of traits [56]. Avery and colleagues adopted a different approach to examining the metabolic syndrome involving the clustering of metabolic traits into six phenotype domains encompassing 19 quantitative traits. Using data from 19,486 European and 6287 African-American individuals, they identified three new loci associated with more than one phenotype domain, APOC1, BRAP and PLCG1 as well as confirming associations of variants near GCKR, ABCB11, LPL, HNF1A, FTO and SUGP1 with multiple phenotype domains [57]. A study in four Finnish cohorts encompassing 11,616 individuals identified a SNP, rs964184, near the APOA1/C3/A4/A5 gene cluster on chromosome 11 as being associated with metabolic syndrome confirming a strong lipid gene component to the genetics of the metabolic syndrome [58]. Three GWAS studies have examined association of genetic variants in non-European populations. A study of 8842 Korean individuals identified two SNPs near BUD13 as reaching genome-wide significance with a further eight SNPs reported to be of nominal significance including previously reported variants near APOA5 and LPL [59]. Metabochip genotype array analysis of an African-American population identified 27 SNPs associated with the metabolic syndrome; however, only one (APOC1) was associated with all five components of the metabolic syndrome [60]. More recently, GWAS on a population of African ancestry identified variants near RALYL, KSR2, MBNL1 and two variants specific to the African population near CA10 and CTNNA3 as being associated with metabolic syndrome [61].
While there is limited overlap between these GWAS, which may, at least in part, be due to slight differences in the definition of metabolic syndrome used, it is notable that many of the identified genes are associated with lipid traits. For example, variants near ZNF259, APOB, LPL, APOA5, CETP and GCKR have been previously shown to be associated with either low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides [62]. Overall, these studies indicate that while there may be a strong lipid element to the genetics underlying the metabolic syndrome, there is no clear evidence at this point in time of one or more pathways linking the different aspects of the syndrome. In a different approach to identify if common metabolic pathways link the various components of the metabolic syndrome, data from 1193 twin men from the Vietnam Era Twin Study of Aging with measures of adiposity (body mass index and waist circumference), blood pressure, insulin resistance (fasting insulin and glucose) and lipids (high-density lipoprotein cholesterol and triglycerides) were analysed [63]. This study suggested that adiposity may be the underlying factor which links the other factors in the metabolic syndrome with findings indicating shared genetic influences between insulin resistance, lipids and adiposity and also between blood pressure and adiposity.
Low-Frequency and Rare Variants
Despite the increased success in identifying risk loci associated with insulin resistance and T2D, it has been estimated that these common variants account for only 25–44 % of the heritability of insulin resistance [64–66]. The contribution of low-frequency (MAF ≤ 5 %) and rare variants (MAF ≤ 0.5 %) to the heritability of T2D and the metabolic syndrome remains to be explored. Exome sequencing, where all protein-coding genes in the genome are sequenced to document all genetic variation in these regions, has recently been employed in an effort to identify novel low-frequency variants that may have large effects on common metabolic phenotypes. Exome sequencing in a Danish cohort of 2000 individuals with a follow-up in a further 15,989 individuals revealed associations of two common SNPs within COBLL1 and MACF1 with T2D and a low-frequency variant in CD300LG with fasting HDL-cholesterol [67]. While the biological function of the variants within COBLL1 and MACF1 is unknown, exploration of the functional effects of the risk variant rs72836561 at CD300LG revealed that this polymorphism is associated with reduced mRNA expression of CD300LG in skeletal muscle and adipose tissue, decreased insulin sensitivity and increased intramyocellular lipid content suggesting a link between this variant and aspects of the metabolic syndrome [68]. Exome sequencing in an Icelandic population revealed that a low-frequency variant in CCND2 reduces the risk of developing T2D but, counterintuitively, is also associated with higher body mass index [69].
The 1000 Genomes Project was established to sequence, through a combination of both genome and exome sequencing, the genomes of 1092 individuals to aid identification of low-frequency and rare genetic variants across populations [70]. These reference panels, containing 38 million SNPs, have been used by the European Network for Genetic and Genomic Epidemiology (ENGAGE) consortium to successfully impute the genotypes of low-frequency variants and perform 22 GWAS for BMI, waist-hip ratio, fasting glucose and fasting insulin. A meta-analysis of these imputed GWAS identified two novel loci for BMI and two novel loci for fasting glucose, of which one was female-specific. The investigators also identified new lead SNPs at 29 established genetic loci including rs1260326 near GCKR for fasting insulin [71].
Another approach to identifying genetic variation associated with complex traits is to examine small populations which have been historically isolated. Such populations are often referred to as founder populations. Genotyping of 2733 individuals from the Greenlandic population using the Cardio-Metabochip followed by exome sequencing identified a common variant within TBC1D4 as being associated with higher fasting glucose and reduced insulin sensitivity [72]. The effect of this p.Arg684Ter variant is to disrupt expression of the full-length TBC1D4 protein in skeletal muscle resulting in decreased insulin-stimulated glucose uptake.
Individuals with familial partial lipodystrophy appear predisposed to features of the metabolic syndrome and insulin resistance. Familial partial lipodystrophy tends to be associated with functional mutations in the LMNA (lamin A/C) gene [73] resulting in altered adipose tissue distribution. Genetic mutations, affecting either LMNA or the associated processing enzyme ZMPSTE24, have been shown to have a prevalence of 3 % in a small cohort of patients with metabolic syndrome [74] raising the possibility that mutations in the LMNA gene may contribute, or predispose, an affected individual to the metabolic syndrome.
Future approaches in identifying low-frequency and rare variants include the UK10K-cohort project which has combined whole-genome sequencing in a cohort of 3781 healthy individuals with exome sequencing of 6000 individuals with either rare disease, severe obesity or a neurodevelopmental disorder [75•]. This project has uncovered 24 million novel genetic variants and resulted in the generation of reference panels with increased coverage of low-frequency and rare variants to help facilitate the identification and contribution of rare variants in health and disease.
Summary and Future Directions
Considerable recent progress has been made in the identification of genetic loci that are associated with insulin resistance and the metabolic syndrome. Some are directly involved in insulin action and glucose metabolism, while further work is required to determine the functional relationships between the genetic variants and insulin action. Nonetheless, the identification of validated variants has been challenging for a number of reasons. First, it is evident that the phenotypes (insulin resistance and metabolic syndrome) are influenced by lifestyle and environmental factors which need to be taken into account when exploring the underlying genetic architecture of these traits. Second, the insulin resistance phenotype itself is difficult to quantify, often relying upon indirect measures such as circulating insulin levels. Furthermore, the phenotype is often limited to whole body measurements with no comprehensive analysis of genetic variants influencing tissue-specific insulin resistance.
Despite these concerns, our appreciation of the genetic basis of insulin resistance and metabolic syndrome has evolved through a number of approaches described in this review. The challenge moving forward is to translate this knowledge into clinical practice to help predict and manage related conditions such as type 2 diabetes and cardiovascular disease.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Alberti KG, Eckel RH, Grundy SM, et al. Harmonizing the metabolic syndrome: a joint interim statement of the international Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120(16):1640–5.
Guallar-Castillon P, Perez RF, Lopez Garcia E, et al. Magnitude and management of metabolic syndrome in Spain in 2008–2010: the ENRICA study. Rev Esp Cardiol. 2014;67(5):367–73.
Prasad DS, Kabir Z, Dash AK, et al. Prevalence and risk factors for metabolic syndrome in Asian Indians: a community study from urban eastern India. J Cardiovasc Dis Res. 2012;3(3):204–11.
Ford ES, Li C, Zhao G, et al. Prevalence of the metabolic syndrome among U.S. adolescents using the definition from the International Diabetes Federation. Diabetes Care. 2008;31(3):587–9.
Sakul H, Pratley R, Cardon L, et al. Familiality of physical and metabolic characteristics that predict the development of non-insulin-dependent diabetes mellitus in Pima Indians. Am J Hum Genet. 1997;60(3):651–6.
Elbein SC, Hasstedt SJ, Wegner K, et al. Heritability of pancreatic beta-cell function among nondiabetic members of Caucasian familial type 2 diabetic kindreds. J Clin Endocrinol Metab. 1999;84(4):1398–403.
Watanabe RM, Valle T, Hauser ER, et al. Familiality of quantitative metabolic traits in Finnish families with non-insulin-dependent diabetes mellitus. Finland-United States Investigation of NIDDM Genetics (FUSION) Study investigators. Hum Hered. 1999;49(3):159–68.
Lehtovirta M, Kaprio J, Forsblom C, et al. Insulin sensitivity and insulin secretion in monozygotic and dizygotic twins. Diabetologia. 2000;43(3):285–93.
Lin HF, Boden-Albala B, Juo SH, et al. Heritabilities of the metabolic syndrome and its components in the northern Manhattan family study. Diabetologia. 2005;48(10):2006–12.
Bellia A, Giardina E, Lauro D, et al. “The linosa study”: epidemiological and heritability data of the metabolic syndrome in a Caucasian genetic isolate. Nutr Metab Cardiovasc Dis. 2009;19(7):455–61.
Carmelli D, Cardon LR, Fabsitz R. Clustering of hypertension, diabetes, and obesity in adult male twins: same genes or same environments? Am J Hum Genet. 1994;55(3):566–73.
Mohlke KL, Boehnke M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum Mol Genet. 2015;24(R1):R85–92.
Knowles JW, Xie W, Zhang Z, et al. Identification and validation of N-acetyltransferase 2 as an insulin sensitivity gene. J Clin Invest. 2015;125(4):1739–51.
Sim E, Abuhammad A, Ryan A. Arylamine N-acetyltransferases: from drug metabolism and pharmacogenetics to drug discovery. Br J Pharmacol. 2014;171(11):2705–25.
Finucane FM, Luan J, Wareham NJ, et al. Correlation of the leptin:adiponectin ratio with measures of insulin resistance in non-diabetic individuals. Diabetologia. 2009;52(11):2345–9.
Pacini G, Mari A. Methods for clinical assessment of insulin sensitivity and beta-cell function. Best Pract Res Clin Endocrinol Metab. 2003;17(3):305–22.
Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–16.
Ingelsson E, Langenberg C, Hivert MF, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59(5):1266–75.
Dimas AS, Lagou V, Barker A, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63(6):2158–71.
Manning AK, Hivert MF, Scott RA, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–69.
Kraft P, Yen YC, Stram DO, et al. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63(2):111–9.
Manning AK, LaValley M, Liu CT, et al. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP x environment regression coefficients. Genet Epidemiol. 2011;35(1):11–8.
Voight BF, Kang HM, Ding J, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793.
Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44(9):991–1005.
Scott RA, Fall T, Pasko D, et al. Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes. 2014;63(12):4378–87. Insulin resistance genetic risk scores were used to highlight that insulin resistance can occur in the absence of obesity.
Yaghootkar H, Scott RA, White CC, et al. Genetic evidence for a normal-weight “metabolically obese” phenotype linking insulin resistance, hypertension, coronary artery disease, and type 2 diabetes. Diabetes. 2014;63(12):4369–77. This paper highlights the relationship between adipose tissue distribution and insulin resistance.
Deeb SS, Fajas L, Nemoto M, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20(3):284–7.
Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26(1):76–80.
Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–89.
Yang M, Ren Y, Lin Z, et al. Kruppel-like factor 14 increases insulin sensitivity through activation of PI3K/Akt signal pathway. Cell Signal. 2015;27(11):2201–8.
Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41(10):1110–5.
Kilpelainen TO, Zillikens MC, Stancakova A, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43(8):753–60.
Van Schaftingen E. A protein from rat liver confers to glucokinase the property of being antagonistically regulated by fructose 6-phosphate and fructose 1-phosphate. Eur J Biochem. 1989;179(1):179–84.
Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94.
Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10(1):51–61.
Do R, Bailey SD, Desbiens K, et al. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec family study. Diabetes. 2008;57(4):1147–50.
Damcott CM, Pollin TI, Reinhart LJ, et al. Polymorphisms in the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in the Amish: replication and evidence for a role in both insulin secretion and insulin resistance. Diabetes. 2006;55(9):2654–9.
Elbein SC, Chu WS, Das SK, et al. Transcription factor 7-like 2 polymorphisms and type 2 diabetes, glucose homeostasis traits and gene expression in US participants of European and African descent. Diabetologia. 2007;50(8):1621–30.
Liu PH, Chang YC, Jiang YD, et al. Genetic variants of TCF7L2 are associated with insulin resistance and related metabolic phenotypes in Taiwanese adolescents and Caucasian young adults. J Clin Endocrinol Metab. 2009;94(9):3575–82.
Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117(8):2155–63.
Shu L, Sauter NS, Schulthess FT, et al. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008;57(3):645–53.
Bailey KA, Savic D, Zielinski M, et al. Evidence of non-pancreatic beta cell-dependent roles of Tcf7l2 in the regulation of glucose metabolism in mice. Hum Mol Genet. 2015;24(6):1646–54.
Tabassum R, Chauhan G, Dwivedi OP, et al. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes. 2013;62(3):977–86.
Succurro E, Andreozzi F, Marini MA, et al. Low plasma insulin-like growth factor-1 levels are associated with reduced insulin sensitivity and increased insulin secretion in nondiabetic subjects. Nutr Metab Cardiovasc Dis. 2009;19(10):713–9.
Mannino GC, Greco A, De Lorenzo C, et al. A fasting insulin-raising allele at IGF1 locus is associated with circulating levels of IGF-1 and insulin sensitivity. PLoS One. 2013;8(12):e85483.
Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40(6):716–8.
Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–75.
Chen G, Bentley A, Adeyemo A, et al. Genome-wide association study identifies novel loci association with fasting insulin and insulin resistance in African Americans. Hum Mol Genet. 2012;21(20):4530–6.
Richards JB, Waterworth D, O’Rahilly S, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5(12):e1000768.
Ruan H, Dong LQ. Adiponectin signaling and function in insulin target tissues. J Mol Cell Biol 2016.
Okada-Iwabu M, Yamauchi T, Iwabu M, et al. A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature. 2013;503(7477):493–9.
Montori-Grau M, Guitart M, Lerin C, et al. Expression and glycogenic effect of glycogen-targeting protein phosphatase 1 regulatory subunit GL in cultured human muscle. Biochem J. 2007;405(1):107–13.
Depetris RS, Hu J, Gimpelevich I, et al. Structural basis for inhibition of the insulin receptor by the adaptor protein Grb14. Mol Cell. 2005;20(2):325–33.
Stancakova A, Laakso M. Genetics of metabolic syndrome. Rev Endocr Metab Disord. 2014;15(4):243–52.
Zabaneh D, Balding DJ. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS One. 2010;5(8):e11961.
Kraja AT, Vaidya D, Pankow JS, et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 2011;60(4):1329–39.
Avery CL, He Q, North KE, et al. A phenomics-based strategy identifies loci on APOC1, BRAP, and PLCG1 associated with metabolic syndrome phenotype domains. PLoS Genet. 2011;7(10):e1002322.
Kristiansson K, Perola M, Tikkanen E, et al. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet. 2012;5(2):242–9.
Jeong SW, Chung M, Park SJ, et al. Genome-wide association study of metabolic syndrome in Koreans. Genom Inform. 2014;12(4):187–94.
Carty CL, Bhattacharjee S, Haessler J, et al. Analysis of metabolic syndrome components in >15 000 African Americans identifies pleiotropic variants: results from the population architecture using genomics and epidemiology study. Circ Cardiovasc Genet. 2014;7(4):505–13.
Tekola-Ayele F, Doumatey AP, Shriner D, et al. Genome-wide association study identifies African-ancestry specific variants for metabolic syndrome. Mol Genet Metab. 2015;116(4):305–13.
Waterworth DM, Ricketts SL, Song K, et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010;30(11):2264–76.
Panizzon MS, Hauger RL, Sailors M, et al. A new look at the genetic and environmental coherence of metabolic syndrome components. Obesity (Silver Spring). 2015;23(12):2499–507.
Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456(7218):18–21.
Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–50.
Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53.
Albrechtsen A, Grarup N, Li Y, et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56(2):298–310.
Stoy J, Kampmann U, Mengel A, et al. Reduced CD300LG mRNA tissue expression, increased intramyocellular lipid content and impaired glucose metabolism in healthy male carriers of Arg82Cys in CD300LG: a novel genometabolic cross-link between CD300LG and common metabolic phenotypes. BMJ Open Diab Res Care. 2015;3(1):e000095.
Steinthorsdottir V, Thorleifsson G, Sulem P, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46(3):294–8.
Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65.
Horikoshi M, Mgi R, van de Bunt M, et al. Discovery and fine-mapping of glycaemic and obesity-related trait loci using high-density imputation. PLoS Genet. 2015;11(7):e1005230.
Moltke I, Grarup N, Jorgensen ME, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512(7513):190–3.
Savage DB, Soos MA, Powlson A, et al. Familial partial lipodystrophy associated with compound heterozygosity for novel mutations in the LMNA gene. Diabetologia. 2004;47(4):753–6.
Dutour A, Roll P, Gaborit B, et al. High prevalence of laminopathies among patients with metabolic syndrome. Hum Mol Genet. 2011;20(19):3779–86.
The UK10K project identifies rare variants in health and disease. Nature 2015. This paper describes the generation of reference panels from the genome and exome sequencing of 10,000 individuals to aid identification of low frequency and rare variants associated with disease.
Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–45.
Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44(9):981–90.
Mahajan A, Go MJ, Zhang W, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–44.
Acknowledgments
Work in Mark Walker’s lab is supported by the National Institute for Health Research (NIHR) Newcastle Biomedical Research Centre based at Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Audrey E. Brown and Mark Walker declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Cardiovascular Genomics
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Brown, A.E., Walker, M. Genetics of Insulin Resistance and the Metabolic Syndrome. Curr Cardiol Rep 18, 75 (2016). https://doi.org/10.1007/s11886-016-0755-4
Published:
DOI: https://doi.org/10.1007/s11886-016-0755-4