Abstract
Purpose of Review
Non-steroidal exacerbated respiratory disease (N-ERD) currently requires aspirin challenge testing for diagnosis. Urinary leukotriene E4 (uLTE4) has been extensively investigated as potential biomarker in N-ERD. We aimed to assess the usefulness of uLTE4 as a biomarker in the diagnosis of N-ERD.
Recent Findings
N-ERD, formerly known as aspirin-intolerant asthma (AIA), is characterised by increased leukotriene production. uLTE4 indicates cysteinyl leukotriene production, and a potential biomarker in N-ERD. Although several studies and have examined the relationship between uLTE4 and N-ERD, the usefulness of uLTE4 as a biomarker in a clinical setting remains unclear.
Findings
Our literature search identified 38 unique eligible studies, 35 were included in the meta-analysis. Meta-analysis was performed (i.e. pooled standardised mean difference (SMD) with 95% confidence intervals (95% CI)) and risk of bias assessed (implementing Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy (Cochrane DTA)). Data from 3376 subjects was analysed (1354 N-ERD, 1420 ATA, and 602 HC). uLTE4 was higher in N-ERD vs ATA (n = 35, SMD 0.80; 95% CI 0.72–0.89). uLTE4 increased following aspirin challenge in N-ERD (n = 12, SMD 0.56; 95% CI 0.26–0.85) but not ATA (n = 8, SMD 0.12; CI − 0.08–0.33). This systematic review and meta-analysis showed that uLTE4 is higher in N-ERD than ATA or HC. Likewise, people with N-ERD have greater increases in uLTE4 following aspirin challenge. However, due to the varied uLTE4 measurement and result reporting practice, clinical utility of these findings is limited. Future studies should be standardised to increase clinical significance and interpretability of the results.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
NSAID-exacerbated respiratory disease (N-ERD) or aspirin exacerbated respiratory disease (AERD), formerly known as aspirin-intolerant asthma (AIA) and Samter’s triad, is a phenotype of asthma characterised by increased leukotriene production and leukotriene driven inflammation [1]. N-ERD is the name used henceforth as it is the term accepted in current clinical practice [2••].
N-ERD is clinically characterised by the presence of asthma, chronic rhinosinusitis with nasal polyposis, and exacerbation of respiratory symptoms on exposure to substances having cyclo-oxygenase 1 (COX-1) inhibiting activity [1, 3•]. The prevalence of N-ERD is reported to be 7% of asthmatics overall and approximately 15% in those who have severe asthma [4]. However, it occurs in 30–40% of those with asthma and nasal polyposis [5]. Accurate diagnosis of this asthma phenotype requires provocation testing, which involves nasal, oral, or inhaled challenge with aspirin [6, 7]. These procedures, whilst being clinically validated, do carry some inherent risks including significant bronchospasm and are thus not recommended for patients with severe airways disease. For these patients, diagnosis of N-ERD has typically relied on medical history alone, which increases the risk of misdiagnosing N-ERD, and the likelihood of providing inappropriate health management, by withholding the use of this class of medication in non-NERD individuals [2••]. Consequently, it is considered highly desirable to identify a robust, accessible, and safe biomarker of N-ERD.
Given that leukotriene status is heightened in N-ERD, there is significant interest in establishing their utility as candidate biomarkers for the diagnosis and disease/treatment monitoring in N-ERD. More specifically, urinary leukotriene E4 (uLTE4) excretion has been identified as a surrogate marker of leukotriene production in vivo and is preferred to other leukotrienes (e.g. Leukotrienes B4, C4, and D4), which have a short half-life and are difficult to measure [8, 9]. To this extent, Hagan et al. [10] reviewed the role of uLTE4 in the diagnosis of N-ERD in 2016. This is the only previous systematic review, of 10 studies, and showed uLTE4 as a biomarker for N-ERD. However, the inclusion criteria for that review [10] required the availability of primary level data to carry out the necessary analysis, and a proportion of full text manuscripts were not available to the authors.
Therefore, in this present study we sought to update the work carried out by Hagan et al. [10], whilst reviewing and analysing the broader literature on this subject to compare the baseline uLTE4 levels in patients with N-ERD, aspirin tolerant asthma (ATA), and healthy control (HC) subjects. In addition, we aimed to determine the impact of aspirin challenge testing on uLTE4 concentration in N-ERD and ATA individuals and the diagnostic accuracy of baseline uLTE4 measurements to predict aspirin intolerance in patients with asthma. In keeping with Hagan et al. [10], we analysed the different assays separately, given the variations in these techniques.
Methods
Literature Search
The protocol for the review was published in the PROSPERO database (CRD42021228674) and developed with reference to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) 2020 guidelines [11]. A systematic search of MEDLINE, EMBASE, EMCARE, CINAHL and PsycINFO was undertaken by a medical librarian in conjunction with one reviewer (B.V.T.) from database inception to 31st December 2021. In contrast to the previous review, a comprehensive search strategy was implemented which captured all studies reporting baseline uLTE4 levels in N-ERD and ATA groups, irrespective of whether these studies reported primary level data to answer our primary research question. No filters were used. The strategies were peer reviewed by a second reviewer (M.M.) prior to final execution of the search. Reference lists from included studies and review articles that were identified through the database searches were hand searched to identify additional articles for possible inclusion. Both Healthcare Databases Advanced Search (HDAS) and Rayyan were used to identify duplicate records and additional duplicates were manually removed before screening for inclusion. Articles were screened by two independent reviewers (B.V.T., M.M.). Disagreements between the reviewers were resolved through discussion. The full search strategy can be found in Online Resource 1.
Study Eligibility
The following medical diagnosis terminologies, i.e. N-ERD/AERD, Samter’s triad, and AIA, have been interchangeably used in the literature to describe the population of interest and were included within the search criteria to ensure completeness of data capture and synthesis.
Original research studies recruiting human subjects with asthma utilising uLTE4 as a biomarker (index test) to differentiate N-ERD from ATA were considered for inclusion. Diagnosis of N-ERD required at least one of the following two criteria to be met (reference standard): (a) positive aspirin challenge, either historic (case–control study design) or performed prospectively (singe-gate design); (b) unequivocal history of asthma exacerbation following ingestion of aspirin and/or other NSAIDs. There were no age restrictions.
The following exclusion criteria were applied: publication types other than primary studies (review articles, case reports, conference abstracts, book chapters and letters to the editor); papers published in languages other than English if a translation could not be found. Studies concerning aspirin challenge testing of asthmatic patients were excluded if baseline (pre-challenge) uLTE4 data was not reported in the published article, in supplementary material, or on request from the corresponding author of the publication.
Study Outcomes
The primary study outcome was to determine whether uLTE4 concentration at baseline in N-ERD is different from ATA and (non-asthmatic) HC subjects, using a between-group comparison. Secondary outcomes were (a) to determine the diagnostic accuracy of baseline uLTE4 measurements to predict aspirin intolerance in patients with asthma; and (b) to determine the change in uLTE4 concentration in N-ERD and ATA following aspirin challenge testing.
Data Extraction
Two reviewers (B.V.T., M.M.) independently extracted the following data from included studies: author(s); year of publication; country of origin; source of funding; demographic characteristics (n, sex, age); clinical characteristics (inclusion/exclusion criteria, co-morbidities, definition of asthma, baseline pulmonary function); index test (method of uLTE4 analysis, original units, nature of urine collection); reference standard (clinical history/aspirin challenge/both, criteria for N-ERD); mean and standard deviation (SD) of uLTE4 at baseline for N-ERD, ATA and HC; diagnostic test accuracy (if reported—area under curve, cut-off value, sensitivity, specificity, positive predictive value, negative predictive value); mean and SD of uLTE4 following aspirin challenge testing for N-ERD and ATA (if performed). Two attempts at requesting missing data from the corresponding authors of included studies were made by contacting them via e-mail. Disagreements in data extraction were resolved through discussion.
If relevant data concerning baseline and/or post-challenge uLTE4 were presented in published figures but not specified as summary data in the accompanying text or supplementary materials, the underlying numerical data was extracted from relevant figures using WebPlotDigitizer (v4.4, California, USA), a web-based semi-automated extraction tool [12].
Risk of Bias Assessment
A modified version of the QUADAS tool from the Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy was used to assess the methodological quality of included studies [13]. This was performed independently by two reviewers (B.V.T., M.M.), with disagreements resolved through discussion.
Data Synthesis and Meta-analysis
A descriptive synthesis of included studies was performed and structured around the review objectives. Studies reporting the mean and SD of uLTE4 at baseline (± post-challenge) for N-ERD, ATA, and HC were included in our meta-analysis. If the extracted data were described as the median with range, or the median with interquartile range, then the data were converted to mean and SD using established approximation methods [14]. Data presented in separate subgroups were combined using established formulae from the Cochrane Handbook for Systematic Reviews of Interventions [15]. Pooled standardised mean difference (SMD) and 95% confidence intervals (CI) were calculated. We investigated the presence of statistical heterogeneity among included studies by using the I2 test. The random-effects model was used if there was significant heterogeneity (I2 > 50%), otherwise the fixed-effects model was used to combine the results. To explore possible sources of heterogeneity, meta-regression analysis was performed, with variables including publication year, country of study origin, sample size, male percentage, and baseline lung function. Any p values of < 0.05 were considered statistically significant.
In a change to the planned data synthesis as registered in PROSPERO, summary receiver-operating characteristic (SROC) modelling was not performed since individual data points were largely missing from included studies. Hence, evaluation of test diagnostic accuracy was not possible.
All data were extracted and stored in an Excel data file (Microsoft Excel for Mac; Microsoft Corporation, USA). Review Manager version 5.4 (The Cochrane Collaboration, Copenhagen, Denmark) and R software version 4.0.1 (R Foundation for Statistical Computing, Vienna, Austria) were used for conducting the meta-analysis.
Results
Study Selection
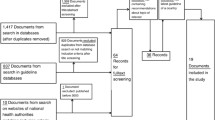
A total of 660 articles were identified [December 2021], with 547 article titles and abstracts reviewed following de-duplication. Of these, 491 articles were ineligible for full-text review. A total of 38 eligible full-text articles were reviewed (Fig. 1). Each article described a unique study. We performed qualitative synthesis of all included studies (n = 38) and meta-analysis of 35 studies. Three of the studies which did not meet the criteria for inclusion in the meta-analysis did not have the required effect size data to allow for such an analysis.
Study Characteristics
Included studies (n = 38) were published between 1991 and 2021, across 8 countries [study numbers as follows: Japan (n = 13), Poland (n = 11), USA (n = 5), South Korea (n = 3), Sweden (n = 2), United Kingdom (n = 2), Italy (n = 1), Switzerland (n = 1)]. A total of n = 1354 N-ERD, n = 1420 ATA, and n = 602 HC subjects were represented across the included studies, with n = 1010 (36.5%) males. In 19 studies, patients with N-ERD were study-defined N-ERD and/or there was clear documentation concerning co-morbid chronic rhinosinusitis and/or nasal polyposis status. In the remaining studies (n = 19), the terminology AIA was used without reference to presence of nasal polyposis. The main characteristics of included studies are summarised in Table 1.
Across all the studies included in this review, uLTE4 concentration was measured using one of 4 different techniques: (i) Amersham-enzyme immunoassay (A-EIA) (n = 8), (ii) Cayman-enzyme immunoassay (C-EIA) (n = 18), (iii) mass spectrometry (MS) (n = 7), and (iv) radioimmunoassay (RIA) (n = 6), with Sanak et al. reporting results with both C-EIA and MS (thus represented twice in these overview data) [16].
Twenty-seven studies used positive aspirin challenge alone (inhaled, intravenous, nasal, or oral) as the reference standard to diagnose N-ERD, two studies used convincing clinical history of asthma exacerbation secondary to ingestion of aspirin alone, and the remaining nine studies used either positive challenge or convincing clinical history. Further details on the aspirin challenge criteria and methodology for uLTE4 measurement are found in Table 2.
Key Findings
Studies with different uLTE4 measurement methodologies were combined. Thirty-five studies including 1127 N-ERD and 1191 ATA reported that the baseline concentration of uLTE4 was significantly higher in N-ERD (SMD 0.80, 95% CI = 0.72 to 0.89; I2 = 42%, Fig. 2) [16–46, 47, 48, 49•, 50•]. Fifteen studies including 780 ATA and 452 HC reported that the baseline concentration of uLTE4 was significantly higher in ATA (SMD 0.45, 95% CI = 0.17 to 0.74; I2 = 78%, Fig. 3) [16, 19, 21–26, 30, 32, 35, 36, 38, 43, 49•]. The concentration of uLTE4 increased following aspirin challenge in N-ERD (12 studies, n = 314 SMD 0.56; 95% CI = 0.26 to 0.85, Fig. 4) [25, 33–35, 37–41, 44, 46, 47] but not ATA (8 studies, n = 187, SMD 0.12; 95% CI = −0.08 to 0.33, Fig. 5) [16, 19, 21–26, 30, 32, 35, 36, 38, 43].
Meta-regression and Risk of Bias
Heterogeneity observed between studies in this meta-analysis was low. Despite this, we performed meta-regression analysis to assess the contribution of several covariates on effect size across studies included in pooling of effect size for baseline uLTE4 in N-ERD vs ATA comparison. I2 for this analysis was low (42%). Meta-regression revealed that country of study had an impact on effect size (I2 = 13.05%). Furthermore, by identifying different study sites and including this in the multiple regression analysis, we found that this would account for an I2 of 100%, suggesting that heterogeneity across studies in this meta-analysis is related to site. There was no significant impact on the effect size when other covariates (publication year, percentage male participants, baseline lung function, and methodology for uLTE4 measurement) were analysed by means of meta-regression, and hence no significant impact on heterogeneity between studies was noted.
Risk of bias assessed by means of the QUADAS tool from the Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy [13], was acceptable across all studies; however 37.8% of quality assessment items were unfulfilled (Figs. 6 and 7). The following risk of bias items were poorly reported across all studies (reported in < 30% overall): spectrum of representative patients (10.5%) and independent interpretation of index and reference standard tests (0%).
Discussion
Our meta-analysis of 35 studies demonstrated a statistically significant higher baseline concentration of uLTE4 in patients with N-ERD compared to those with ATA and HC, adding an addition 25 studies to the previous review. These findings corroborate current knowledge regarding the importance of leukotriene status in patients with N-ERD, and again identify uLTE4 as a potential biomarker in N-ERD diagnosis and disease monitoring. For the subset of studies reporting uLTE4 measurements before and after aspirin challenge testing, a significant rise in uLTE4 was seen in patients with N-ERD, but not those with ATA. This is the first meta-analysis which evaluates the change in uLTE4 concentrations following aspirin challenge in N-ERD compared to ATA, and the results are consistent with previous literature demonstrating that the magnitude of nasal and/or respiratory reactions to provocative aspirin challenges in asthmatics is associated with both the degree of baseline uLTE4 elevation and the rise in uLTE4 during a challenge [51, 52].
This study has a number of limitations. Because individual data points were largely missing from most studies, sensitivity and specificity testing was not possible. Four studies did provide some data of interest [8, 9, 16, 38], but this was insufficient to carry out this analysis. The corresponding authors of the rest of the included studies were contacted via e-mail asking for this data, but there was no response from any of them. Studies included were published between 1991 to 2021, a total span of 30 years, and this will invariably carry with it a variation in practice of uLTE4 measurement. Although, our meta-regression analysis did not identify year of publication as contributing to heterogeneity across studies, four different methodologies were used to measure uLTE4 across the studies included. However, to account for this, a separate comparison analysis for studies using each of the methods was performed and then the studies were combined. This analysis has revealed that despite the different methodologies, there was no significant heterogeneity across studies (Fig. 2), meaning that different methodologies were not shown to have a significant impact on effect size. Although the different methodologies did not appear to result in heterogeneity, there was a large number of methodologies used and methods of reporting the data. The country of publication had an effect on heterogeneity but not when site was included in the multiple regression. This suggests that site was responsible for the heterogeneity, presumably due to a composite of methodology, definition of N-ERD and population sampled. Greater standardisation of the procedure and reporting is required in clinical research and clinical practice.
There was also variation in the way asthma was defined across studies, with American Thoracic Society (ATS) criteria, Global Initiative for Asthma (GINA) guidelines, National Heart, Lung and Blood Institute criteria, and physician diagnosis all used. In 17 studies, definition of asthma was not specified. This is important given that it will dictate the characteristics of the population being studied. Similarly, the definition of aspirin intolerance varied across studies. Although most studies performed aspirin challenge testing (either retrospectively or prospectively), there was considerable variation in the challenge agent employed and the diagnostic cut-off for a positive test (i.e., fall in FEV1 relative to baseline). Approximately half of studies included in the meta-analysis (18/35) provided clear documentation of co-morbid chronic rhinosinusitis and/or nasal polyposis status, or the aspirin-intolerant cohort was defined as N-ERD. The remaining studies did not provide such population characteristics. In several studies, summary data concerning uLTE4 levels were not stated in the published text or supplementary materials and had to be derived from figures using a web-based extraction tool. This invariably is an estimation of the data. Similarly, for studies where the reported data was described as median with range or interquartile range, this required conversion to mean and SD using published approximation methods. This is important because of the potential impact this has on the accuracy of the results and the impact this could have on the weight of the individual studies, and therefore the overall study results. We therefore feel that standardisation of result reporting should also be implemented.
One of the most important features of this meta-analysis is the enforced use of the standardised mean difference. This summary statistic is used when the measurement scales of the various papers are too diverse to be pooled in a meta-analysis, and thus they have to be converted to a common statistical denominator, or statistical units. The use of the standardised difference means that we cannot know the absolute difference between groups, nor can we define a diagnostic cut off. This is important especially when considering developing study protocols going forward with the aim of establishing sensitivity and specificity. This work has identified the need for standardisation of such protocols to move closer towards achieving clinical significance. Our results show that all the methodologies employed to measure uLTE4 yielded comparable results across studies. Mass spectrometry has been described in a number of publications as the gold standard for the measurement of leukotrienes in biological fluids [53, 54]; however, access to MS and cost might impact its availability in the clinical setting, whereas, enzyme immunoassays might be more readily available. We feel that these are important considerations to make going forward in the protocol development for research of this subject area. This would allow calculation of the absolute mean difference in clinically useful terms rather than the slightly abstract concept of a standardised mean difference. The current heterogeneity in methods and measurement makes it impossible to come up with clinically relevant recommendations on the use of such diagnostic technology.
It should also be noted that most studies have been conducted in specialist centres and excluded participants with uncontrolled asthma or participants reporting a respiratory tract infection or asthma exacerbation in the preceding 6 weeks. While this provides a well-defined cohort for research purposes, our findings may not be generalisable to patients undergoing testing in routine clinical practice, especially since N-ERD is most prevalent among patients with severe asthma.
Overall, the risk of bias was acceptable across all studies. However, in all included studies, it was not reported whether study authors were blinded to baseline uLTE4 data (index test) when performing aspirin challenge testing or obtaining clinical history of aspirin intolerance (reference standard). The primary aim of many included studies was not to determine test diagnostic accuracy, which may account for this. It is also unclear how much a lack of blinding could affect interpretation of aspirin challenge testing since challenges are normally undertaken following a set protocol with a pre-determined diagnostic cut-off.
The finding of a significant rise in uLTE4 following aspirin challenge testing is in keeping with the central role leukotriene release as a cause of upper and lower airway symptoms [44]. Daffern et al. showed that rise in uLTE4 following challenge was related to severity of airflow obstruction post challenge. However interestingly the rise does not seem to be attenuated by inhibition of 5-lipoxygenase which should reduce leukotriene production [51, 55].
Conclusion
The true prevalence of N-ERD is unclear and it is likely to be significantly underdiagnosed especially in those individuals with mild respiratory symptoms, and because of difficulty accessing specialist centres for diagnostic confirmation [2••, 4]. An accurate diagnosis of N-ERD is important, as this can have an impact on both treatment modalities and management of co-morbid chronic diseases such as ischaemic heart disease and chronic pain. Including uLTE4 in the diagnostic algorithm for patients suspected to suffer from N-ERD would be especially useful in individuals who may be at higher risk of adverse reactions from aspirin challenge testing because of increased risk such as FEV1 < 70%, or nasal pathology (precluding nasal aspirin challenge test) [2••]. This safe, non-invasive biomarker for N-ERD may reduce clinician time needed for aspirin challenge testing and would be cost-effective. Future research should be directed at evaluating diagnostic specificity and sensitivity to establish biomarker diagnostic accuracy and employing standardised methods of uLTE4 measurements to ensure any results yielded are more readily translatable to impact clinical practice.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
White AA, Stevenson DD. Aspirin-exacerbated respiratory disease. N Engl J Med. 2018;379:1060–70.
•• Kowalski ML, Agache I, Bavbek S, Bakirtas A, Blanca M, Bochenek G, et al. Diagnosis and management of NSAID-exacerbated respiratory disease (N-ERD)-a EAACI position paper. Allergy. 2019;74:28–39. Position paper summarising current knowledge on the pathophysiology of N-ERD, existing diagnostic tools, and future directions for research.
• Wangberg H, White AA. Aspirin-exacerbated respiratory disease. Curr Opin Immunol. 2020;66:9–13. Review detailing current understanding of the immunopathogenesis of N-ERD.
Rajan JP, Wineinger NE, Stevenson DD, White AA. Prevalence of aspirin-exacerbated respiratory disease among asthmatic patients: a meta-analysis of the literature. J Allergy Clin Immunol. 2015;135:676-681.e1.
Jenkins C, Costello J, Hodge L. Systematic review of prevalence of aspirin induced asthma and its implications for clinical practice. BMJ. 2004;328:434.
Laidlaw TM. Clinical updates in aspirin-exacerbated respiratory disease. Allergy Asthma Proc. 2019;40:4–6.
Miller B, Mirakian R, Gane S, Larco J, Sannah AA, Darby Y, et al. Nasal lysine aspirin challenge in the diagnosis of aspirin - exacerbated respiratory disease: asthma and rhinitis. Clin Exp Allergy. 2013;43:874–80.
Bochenek G, Stachura T, Plutecka H, Sanak M, Nizankowska-Mogilnicka E, Sladek K, et al. Diagnostic accuracy of urinary LTE4 measurement to predict aspirin-exacerbated respiratory disease in patients with asthma. J Allerg Clin Immunol: In Practice. Ame Acad Allerg Asthma Immun. 2018;6:528–35.
Comhair SAA, Bochenek G, Baicker-McKee S, Wang Z, Stachura T, Sanak M, et al. The utility of biomarkers in diagnosis of aspirin exacerbated respiratory disease. Respir Res. 2018;19:210.
Hagan JB, Laidlaw TM, Divekar R, O’Brien EK, Kita H, Volcheck GW, et al. Urinary leukotriene E4 to determine aspirin intolerance in asthma: a systematic review and meta-analysis. J Allergy Clin Immunol Pract. 2017;5:990–990.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372: n71.
Drevon D, Fursa SR, Malcolm AL. Intercoder reliability and validity of WebPlotDigitizer in extracting graphed data. Behav Modif. 2017;41:323–39.
Reitsma JB, Rutjes A, Whiting P, Vlassov V, Leeflang M, Deeks J. Chapter 9: Assessing methodological quality. Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy. 2009.
Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135.
Higgins J, Abe S. Cochrane handbook for systematic reviews of interventions version 5.1.0. The Cochrane Collab. 2011. Available from: https://www.handbook.cochrane.org.
Sanak M, Bochenek G, Faber J, Plutecka H, Szczeklik A. Elevated urinary leukotriene E excretion in asthma: a comparison of HPLC-mass spectrometry and ELISA. Allergy. 2010;65:663–4.
Higashi N, Taniguchi M, Mita H, Osame M, Akiyama K. A comparative study of eicosanoid concentrations in sputum and urine in patients with aspirin-intolerant asthma. Clinical and experimental allergy : J Br Soc Allerg Clin Immunol. 2002;32:1484–90.
Higashi N, Taniguchi M, Mita H, Ishii T, Akiyama K. Nasal blockage and urinary leukotriene E4 concentration in patients with seasonal allergic rhinitis. Allergy. 2003;58:476–80.
Kawagishi Y, Mita H, Taniguchi M, Maruyama M, Oosaki R, Higashi N, et al. Leukotriene C4 synthase promoter polymorphism in Japanese patients with aspirin-induced asthma. J Allergy Clin Immunol. 2002;109:936–42.
Mita H, Endoh S, Kudoh M, Kawagishi Y, Kobayashi M, Taniguchi M, et al. Possible involvement of mast-cell activation in aspirin provocation of aspirin-induced asthma. Allergy. 2001;56:1061–7.
Mitsui C, Kajiwara K, Hayashi H, Kinoshita A, Fukutomi Y, Sekiya K, et al. Platelet activation markers overexpressed specifically in aspirinexacerbated respiratory disease. Allergy: European J Allerg Clin Immun. 2015;70:636–636.
Oosaki R, Mizushima Y, Kawasaki A, Kashii T, Mita H, Shida T, et al. Urinary excretion of leukotriene E4 and 11-dehydrothromboxane B2 in patients with spontaneous asthma attacks. Int Arch Allergy Immunol. 1997;114:373–8.
Yamaguchi H, Higashi N, Mita H, Ono E, Komase Y, Nakagawa T, et al. Urinary concentrations of 15-epimer of lipoxin A(4) are lower in patients with aspirin-intolerant compared with aspirin-tolerant asthma. Clinical and experimental allergy : J Br Soc Allerg Clin Immunol. 2011;41:1711–8.
Yamaguchi, Ishii T, Yamamoto K, Higashi N, Taniguchi M, Okamoto M. Differences in urinary leukotriene E4 levels and distribution of eosinophils between chronic rhinosinusitis patients with aspirin-intolerant and -tolerant asthma. Auris Nasus Larynx. 2016;43:304–8.
Bochenek G, Nagraba K, Nizankowska E, Szczeklik A. A controlled study of 9alpha,11beta-PGF2 (a prostaglandin D2 metabolite) in plasma and urine of patients with bronchial asthma and healthy controls after aspirin challenge. J Allergy Clin Immunol. 2003;111:743–9.
Bochenek G, Stachura T, Plutecka H, Sanak M, Nizankowska-Mogilnicka E, Sladek K, et al. Diagnostic Accuracy of Urinary LTE4 Measurement to Predict Aspirin-Exacerbated Respiratory Disease in Patients with Asthma. J Allerg Clin Immunol: In Practice. 2018;6:528–35.
Gaber F, Daham K, Higashi A, Higashi N, Gülich A, Delin I, et al. Increased levels of cysteinyl-leukotrienes in saliva, induced sputum, urine and blood from patients with aspirin-intolerant asthma. Thorax. 2008;63:1076–82.
Higashi N, Mita H, Ono E, Fukutomi Y, Yamaguchi H, Kajiwara K, et al. Profile of eicosanoid generation in aspirin-intolerant asthma and anaphylaxis assessed by new biomarkers. J Allergy Clin Immunol. 2010;125:1084–1084.
Jerschow E, Ren Z, Hudes G, Sanak M, Morales E, Schuster V, et al. Utility of low-dose oral aspirin challenges for diagnosis of aspirin-exacerbated respiratory disease. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2016;116:321–321.
Mastalerz L, Sanak M, Szczeklik A. Serum interleukin-5 in aspirin-induced asthma. Clinical and experimental allergy : J Br Soc Allerg Clin Immunol. 2001;31:1036–40.
Mastalerz L, Gawlewicz-Mroczka A, Nizankowska E, Cmiel A, Szczeklik A. Protection against exercise-induced bronchoconstriction by montelukast in aspirin-sensitive and aspirin-tolerant patients with asthma. Clinical and experimental allergy : J Br Soc Allerg Clin Immunol. 2002a;32:1360–5.
Mastalerz L, Sanak M, Gawlewicz-Mroczka A, Gielicz A, Szczeklik A, Cmiel A. Prostaglandin E2 systemic production in patients with asthma with and without aspirin hypersensitivity. Thorax. 2008;63:27–34.
Mastalerz L, Januszek R, Kaszuba M, Wójcik K, Celejewska-Wójcik N, Gielicz A, et al. Aspirin provocation increases 8-iso-PGE2 in exhaled breath condensate of aspirin-hypersensitive asthmatics. Prostaglandins Other Lipid Mediat. 2015;121:163–9.
Micheletto C, Tognella S, Visconti M, Trevisan F, Dal Negro RW. Changes in urinary LTE4 and nasal functions following nasal provocation test with ASA in ASA-tolerant and -intolerant asthmatics. Respir Med. 2006;100:2144–50.
Mita H, Higashi N, Taniguchi M, Higashi A, Akiyama K. Increase in urinary leukotriene B4 glucuronide concentration in patients with aspirin-intolerant asthma after intravenous aspirin challenge. Clinical and experimental allergy : J Br Soc Allerg Clin Immunol. 2004;34:1262–9.
Ono E, Taniguchi M, Higashi N, Mita H, Yamaguchi H, Tatsuno S, et al. Increase in salivary cysteinyl-leukotriene concentration in patients with aspirin-intolerant asthma. Allergology international : official journal of the Japanese Society of Allergology. 2011;60:37–43.
Pezato R, Świerczyńska-Krępa M, Niżankowska-Mogilnicka E, Holtappels G, De Ruyck N, Sanak M, et al. Systemic expression of inflammatory mediators in patients with chronic rhinosinusitis and nasal polyps with and without Aspirin Exacerbated Respiratory Disease. Cytokine. 2016;77:157–67.
Sanak M, Kiełbasa B, Bochenek G, Szczeklik A. Exhaled eicosanoids following oral aspirin challenge in asthmatic patients. Clinical and experimental allergy: J Br Soc Allerg Clin Immunol. 2004;34:1899–904.
Ban GY, S.-H. K, Yoon MG, Kim JH, Shin YS, Ye YM, et al. Serum LTE4 metabolite as a biomarker for aspirin exacerbated respiratory disease. World Allerg Org J. 2016;9:21–21.
Świerczyńska-Krępa M, Sanak M, Bochenek G, Stręk P, Ćmiel A, Gielicz A, et al. Aspirin desensitization in patients with aspirin-induced and aspirin-tolerant asthma: a double-blind study. J Allergy Clin Immunol. 2014;134:883–90.
Cahill KN, Bensko JC, Boyce JA, Laidlaw TM. Prostaglandin D2: a dominant mediator of aspirin-exacerbated respiratory disease. J Allergy Clin Immunol. 2015;135:245–52.
Cahill KN, Cui J, Kothari P, Murphy K, Raby BA, Singer J, et al. Unique effect of aspirin therapy on biomarkers in aspirin-exacerbated respiratory disease. A Prospective Trial. Am J Respir Crit Care Med. 2019;200:704–11.
Laidlaw TM, Kidder MS, Bhattacharyya N, Boyce JA, Milne GL. Increased platelet adherence to leukocytes results in cysteinyl leukotriene (cysLT) overproduction in aspirin exacerbated respiratory disease (AERD). J Allerg Clin Immuno. 2012;129. Available from: https://go.openathens.net/redirector/nhs?url=https%3A%2F%2Fwww.clinicalkey.com%2Fcontent%2FplayBy%2Fdoi%2F%3Fv%3D10.1016%2Fj.jaci.2011.12.455.
Christie PE, Tagari P, Ford-Hutchinson AW, Charlesson S, Chee P, Arm JP, et al. Urinary leukotriene E4 concentrations increase after aspirin challenge in aspirin-sensitive asthmatic subjects. Am Rev Respir Dis. 1991;143:1025–9.
Kumlin M, Dahlén B, Björck T, Zetterström O, Granström E, Dahlén SE. Urinary excretion of leukotriene E4 and 11-dehydro-thromboxane B2 in response to bronchial provocations with allergen, aspirin, leukotriene D4, and histamine in asthmatics. Am Rev Respir Dis. 1992;146:96–103.
Obase Y, Shimoda T, S.-Y. T, Mitsuta K, Fukushima C, Kawano T, et al. Effects of pranlukast on aspirin-induced bronchoconstriction: differences in chemical mediators between aspirin-intolerant and tolerant asthmatic patients. Annals of Allergy, Asthma and Immunology. 2001;87:74–9.
Obase Y, Shimoda T, Tomari S, Mitsuta K, Kawano T, Matsuse H, et al. Effects of pranlukast on chemical mediators in induced sputum on provocation tests in atopic and aspirin-intolerant asthmatic patients. Chest. 2002;121:143–50.
Mastalerz L, Nizankowska E, Sanak M, Mejza F, Pierzchalska M, Bazan-Socha S, et al. Clinical and genetic features underlying the response of patients with bronchial asthma to treatment with a leukotriene receptor antagonist. Eur J Clin Invest. 2002b;32:949–55.
• Ban G-Y, Kim S-H, Park H-S. Persistent eosinophilic inflammation in adult asthmatics with high serum and urine levels of leukotriene E4. J Asthma Allerg. 2021;14:1219–30. Urinary leukotriene E4 remains significantly higher in N-ERD than aspirin-tolerant asthmatics despite leukotriene receptor antagonist treatment.
• Choi Y., Sim S., Lee D.-H., Shin Y.S., Park H.-S., Lee H.-R., et al. Effect of TGF-beta1 on eosinophils to induce cysteinyl leukotriene E4 production in aspirin-exacerbated respiratory disease. PLoS ONE. Pub Lib Sci. 2021;16. Higher levels of TGF-β1 in N-ERD patients may contribute to urinary leukotriene E4 production via enhancing LTC4S expression which induces eosinophil degranulation, accelerating airway inflammation.
Daffern PJ, Muilenburg D, Hugli TE, Stevenson DD. Association of urinary leukotriene E4 excretion during aspirin challenges with severity of respiratory responses. J Allergy Clin Immunol. 1999;104:559–64.
Swierczynska M, Nizankowska-Mogilnicka E, Zarychta J, Gielicz A, Szczeklik A. Nasal versus bronchial and nasal response to oral aspirin challenge: clinical and biochemical differences between patients with aspirin-induced asthma/rhinitis. J Allergy Clin Immunol. 2003;112:995–1001.
Montuschi P, Santini G, Valente S, Mondino C, Macagno F, Cattani P, et al. Liquid chromatography-mass spectrometry measurement of leukotrienes in asthma and other respiratory diseases. J Chromatogr, B: Anal Technol Biomed Life Sci. 2014;964:12–25.
Bochenek G, Niżankowska E, Gielicz A, Świerczyńska M, Szczeklik A. Plasma 9α,11β-PGF2, a PGD2 metabolite, as a sensitive marker of mast cell activation by allergen in bronchial asthma. Thorax BMJ Publishing Group Ltd. 2004;59:459–64.
Pauls JD, Simon RA, Daffern PJ, Stevenson DD. Lack of effect of the 5-lipoxygenase inhibitor zileuton in blocking oral aspirin challenges in aspirin-sensitive asthmatics. Ann Allergy Asthma Immunol. 2000;85:40–5.
Smith CM, Hawksworth RJ, Thien FC, Christi e PE, Lee TH. Urinary leukotriene E4 in bronchial asthma. Eur Respir J. 1992;5(6):693–9.
Funding
This work was funded by Asthma UK Centre of Applied Research (AUKCAR).
Author information
Authors and Affiliations
Contributions
All authors substantially contributed to conception and design of the study, acquisition of the data, or analysis and interpretation of the data; drafted the article or revised it for important intellectual content; gave final approval of the version to be submitted; and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Asthma
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marquette, M., Tailor, B.V., Calder, P.C. et al. Urinary Leukotriene E4 as a Biomarker in NSAID-Exacerbated Respiratory Disease (N-ERD): a Systematic Review and Meta-analysis. Curr Allergy Asthma Rep 22, 209–229 (2022). https://doi.org/10.1007/s11882-022-01049-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11882-022-01049-8