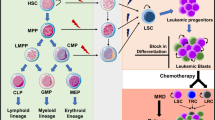
Opinion statement
Acute myeloid leukemia (AML) is the most common form of leukemia in adults, leading to the highest number of annual leukemia-associated deaths in the USA. Although most AML patients initially enter remission following induction therapy, most eventually relapse, underscoring the unmet need for more effective therapies. In recent years, novel high-throughput sequencing techniques, and mouse and human models of disease have increased our understanding of the molecular mechanisms that lead to AML. Leukemogenic mechanisms can be broadly classified into two types—cell-intrinsic and cell-extrinsic. Cell-intrinsic mechanisms include an array of genetic and epigenetic alterations that lead to dysregulated gene expression and function in hematopoietic stem/progenitor cells, leading to their increased fitness and ultimately, malignant transformation. Extrinsic mechanisms include both hematopoietic and non-hematopoietic stromal components of the leukemic microenvironment that interact with pre-leukemic and leukemic clones to promote their survival, self-renewal, and/or resistance to therapy. Through the individual and concerted action of these factors, pre-leukemic clones acquire the changes necessary for leukemic transformation. In addition, following therapy, specific leukemic clones are selected for that eventually re-initiate disease. Improving our understanding of these cell-intrinsic and cell-extrinsic mechanisms will provide novel opportunities to treat AML as well as prevent the development of disease.

Similar content being viewed by others
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–52.
Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–47.
Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–66.
Porter CC. Germ line mutations associated with leukemias. Hematology Am Soc Hematol Educ Program. 2016;2016(1):302–8.
Sison EA, Brown P. The bone marrow microenvironment and leukemia: biology and therapeutic targeting. Expert Rev Hematol. 2011;4(3):271–83.
DiNardo CD, Cortes JE. Mutations in AML: prognostic and therapeutic implications. Hematology Am Soc Hematol Educ Program. 2016;2016(1):348–55.
Frohling S, Scholl C, Gilliland DG, Levine RL. Genetics of myeloid malignancies: pathogenetic and clinical implications. J Clin Oncol. 2005;23(26):6285–95.
Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–21.
Weissman I. Stem cell research: paths to cancer therapies and regenerative medicine. JAMA. 2005;294(11):1359–66.
Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118.
Corces MR, Buenrostro JD, Wu B, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48(10):1193–203.
Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44(1):23–31.
Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24.
Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–9.
Avagyan S, Henninger JE, Mannherz WP, et al. Resistance to inflammation underlies enhanced fitness in clonal hematopoiesis. Science. 2021;374(6568):768–772. Demonstrates inflammation as a critical driver of clonal fitness and survival.
King KY, Goodell MA. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol. 2011;11(10):685–92.
Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366(6465). Excellent summary of the molecular mechanisms driving clonal hematopoiesis.
Steensma DP. Clinical consequences of clonal hematopoiesis of indeterminate potential. Blood Adv. 2018;2(22):3404–3410. Excellent summary of the causes and clinical consequences of clonal hematopoiesis.
Bouzid H, Belk J, Jan M, Qi Y, Sarnowski C, Wirth S, Ma L, Chrostek M, Ahmad H, Nachun D, Yao W, Beiser A, Bick AG, Bis J, Fornage M, Longstreth WT, Lopez O, Nataranjan P, Psaty B, Satizabal C, Weinstock J, Larson E, Crane P, Keene CD, Seshadri S, Satpathy AT, Montine T, Jaiswal S. Clonal hematopoiesis is associated with reduced risk of Alzheimer’s disease. Blood. 2021; (1): 5.
Chen J, Kao YR, Sun D, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med. 2019;25(1):103–110. Demonstrates parallel clonal evolution of mutant MDS clones into leukemic clones.
Strickland SA, Vey N. Diagnosis and treatment of therapy-related acute myeloid leukemia. Crit Rev Oncol Hematol. 2022;171:103607.
Sperling AS, Guerra VA, Kennedy JA, et al. Lenalidomide promotes the development of TP53-mutated therapy-related myeloid neoplasms. Blood. 2022. https://doi.org/10.1182/blood.2021014956. Elucidates a therapy-specific molecular mechanism driving clonal selection and development of therapy-related AML/MDS.
Lindsley RC, Mar BG, Mazzola E, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125(9):1367–76.
Weinberg OK, Siddon A, Madanat YF, et al. TP53 mutation defines a unique subgroup within complex karyotype de novo and therapy-related MDS/AML. Blood Adv. 2022;6(9):2847–2853. Highlights the need to revise the classification scheme of myeloid malignancies for better clinical management of disease.
Roberts I, Izraeli S. Haematopoietic development and leukaemia in Down syndrome. Br J Haematol. 2014;167(5):587–99.
Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549–57.
Niemeyer CM. RAS diseases in children. Haematologica. 2014;99(11):1653–62.
Chang TY, Dvorak CC, Loh ML. Bedside to bench in juvenile myelomonocytic leukemia: insights into leukemogenesis from a rare pediatric leukemia. Blood. 2014;124(16):2487–97.
Spinner MA, Sanchez LA, Hsu AP, et al. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood. 2014;123(6):809–21.
Latger-Cannard V, Philippe C, Bouquet A, et al. Haematological spectrum and genotype-phenotype correlations in nine unrelated families with RUNX1 mutations from the French network on inherited platelet disorders. Orphanet J Rare Dis. 2016;11:49.
Churpek JE, Garcia JS, Madzo J, Jackson SA, Onel K, Godley LA. Identification and molecular characterization of a novel 3′ mutation in RUNX1 in a family with familial platelet disorder. Leuk Lymphoma. 2010;51(10):1931–5.
Noris P, Perrotta S, Seri M, et al. Mutations in ANKRD26 are responsible for a frequent form of inherited thrombocytopenia: analysis of 78 patients from 21 families. Blood. 2011;117(24):6673–80.
Zhang MY, Churpek JE, Keel SB, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47(2):180–5.
Moriyama T, Metzger ML, Wu G, et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: a systematic genetic study. Lancet Oncol. 2015;16(16):1659–66.
Tawana K, Wang J, Renneville A, et al. Disease evolution and outcomes in familial AML with germline CEBPA mutations. Blood. 2015;126(10):1214–23.
Cardoso SR, Ryan G, Walne AJ, et al. Germline heterozygous DDX41 variants in a subset of familial myelodysplasia and acute myeloid leukemia. Leukemia. 2016;30(10):2083–6.
McBride KA, Ballinger ML, Killick E, et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol. 2014;11(5):260–71.
Meyer C, Burmeister T, Groger D, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32(2):273–84.
Giles RH, Dauwerse JG, Higgins C, et al. Detection of CBP rearrangements in acute myelogenous leukemia with t(8;16). Leukemia. 1997;11(12):2087–96.
Friedman AD. Leukemogenesis by CBF oncoproteins. Leukemia. 1999;13(12):1932–42.
Castilla LH, Perrat P, Martinez NJ, et al. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proc Natl Acad Sci U S A. 2004;101(14):4924–9.
Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–33.
Russler-Germain DA, Spencer DH, Young MA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25(4):442–54.
Lu R, Wang P, Parton T, et al. Epigenetic perturbations by Arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell. 2016;30(1):92–107.
Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472–8.
Guryanova OA, Shank K, Spitzer B, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22(12):1488–95.
Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30.
Weissmann S, Alpermann T, Grossmann V, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26(5):934–42.
Shih AH, Jiang Y, Meydan C, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27(4):502–15.
Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27.
Cierpicki T, Risner LE, Grembecka J, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010;17(1):62–8.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
Khalaj M, Woolthuis CM, Hu W, et al. miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J Exp Med. 2017;214(8):2453–70.
Hu W, Dooley J, Chung SS, et al. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood. 2015;125(14):2206–16.
Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111(10):5078–85.
Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16(1):49–58.
Chen P, Price C, Li Z, et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci U S A. 2013;110(28):11511–6.
Senyuk V, Zhang Y, Liu Y, et al. Critical role of miR-9 in myelopoiesis and EVI1-induced leukemogenesis. Proc Natl Acad Sci U S A. 2013;110(14):5594–9.
So AY, Sookram R, Chaudhuri AA, et al. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood. 2014;124(9):1502–12.
Narayan N, Morenos L, Phipson B, et al. Functionally distinct roles for different miR-155 expression levels through contrasting effects on gene expression, in acute myeloid leukaemia. Leukemia. 2017;31(4):808–20.
Wallace JA, Kagele DA, Eiring AM, et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood. 2017;129(23):3074–86.
Huan J, Hornick NI, Goloviznina NA, et al. Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia. 2015;29(12):2285–95.
Hornick NI, Doron B, Abdelhamed S, et al. AML suppresses hematopoiesis by releasing exosomes that contain microRNAs targeting c-MYB. Sci Signal. 2016;9(444):ra88.
Nowek K, Sun SM, Dijkstra MK, et al. Expression of a passenger miR-9* predicts favorable outcome in adults with acute myeloid leukemia less than 60 years of age. Leukemia. 2016;30(2):303–9.
Mishra S, Liu J, Chai L, Tenen DG. Diverse functions of long noncoding RNAs in acute myeloid leukemia: emerging roles in pathophysiology, prognosis, and treatment resistance. Curr Opin Hematol. 2022;29(1):34–43. Up-to-date summary of the role of lncRNA in myeloid malignancies.
Zhang YY, Huang SH, Zhou HR, Chen CJ, Tian LH, Shen JZ. Role of HOTAIR in the diagnosis and prognosis of acute leukemia. Oncol Rep. 2016;36(6):3113–22.
Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11(6):777–87.
Wang H, Li W, Guo R, et al. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int J Cancer. 2014;135(12):2783–94.
Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39(4):330–6.
Zhao TF, Jia HZ, Zhang ZZ, et al. LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol Med Rep. 2017;16(3):3687–93.
Tikhonova AN, Dolgalev I, Hu H, et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569(7755):222–228. Single-cell level mapping of the bone marrow microenvironment and factors regulating hematopoiesis.
Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5(4):311–21.
Poulos MG, Gars EJ, Gutkin MC, et al. Activation of the vascular niche supports leukemic progression and resistance to chemotherapy. Exp Hematol. 2014;42(11):976-986 e973.
Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464(7290):852–7.
Kim YW, Koo BK, Jeong HW, et al. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood. 2008;112(12):4628–38.
Zimmer SN, Zhou Q, Zhou T, et al. Crebbp haploinsufficiency in mice alters the bone marrow microenvironment, leading to loss of stem cells and excessive myelopoiesis. Blood. 2011;118(1):69–79.
Franceschi C, Bonafe M, Valensin S, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–54.
Cai Z, Kotzin JJ, Ramdas B, et al. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell. 2018;23(6):833-849 e835.
Schepers K, Pietras EM, Reynaud D, et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 2013;13(3):285–99.
Medyouf H, Mossner M, Jann JC, et al. Myelodysplastic cells in patients reprogram mesenchymal stromal cells to establish a transplantable stem cell niche disease unit. Cell Stem Cell. 2014;14(6):824–37.
Hanoun M, Zhang D, Mizoguchi T, et al. Acute myelogenous leukemia-induced sympathetic neuropathy promotes malignancy in an altered hematopoietic stem cell niche. Cell Stem Cell. 2014;15(3):365–75.
Muntion S, Ramos TL, Diez-Campelo M, et al. Microvesicles from mesenchymal stromal cells are involved in HPC-microenvironment crosstalk in myelodysplastic patients. PLoS ONE. 2016;11(2):e0146722.
Dias S, Hattori K, Heissig B, et al. Inhibition of both paracrine and autocrine VEGF/ VEGFR-2 signaling pathways is essential to induce long-term remission of xenotransplanted human leukemias. Proc Natl Acad Sci U S A. 2001;98(19):10857–62.
Hussong JW, Rodgers GM, Shami PJ. Evidence of increased angiogenesis in patients with acute myeloid leukemia. Blood. 2000;95(1):309–13.
Ben-Batalla I, Schultze A, Wroblewski M, et al. Axl, a prognostic and therapeutic target in acute myeloid leukemia mediates paracrine crosstalk of leukemia cells with bone marrow stroma. Blood. 2013;122(14):2443–52.
Lane SW, Wang YJ, Lo Celso C, et al. Differential niche and Wnt requirements during acute myeloid leukemia progression. Blood. 2011;118(10):2849–56.
Farge T, Saland E, de Toni F, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7(7):716–35.
Ye H, Adane B, Khan N, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell. 2016;19(1):23–37.
Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315–21.
Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167–74.
Jacamo R, Chen Y, Wang Z, et al. Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of NF-kappaB mediates chemoresistance. Blood. 2014;123(17):2691–702.
Nervi B, Ramirez P, Rettig MP, et al. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113(24):6206–14.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors do not have any potential conflicts of interest to disclose.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Leukemia
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chakraborty, S., Park, C.Y. Pathogenic Mechanisms in Acute Myeloid Leukemia. Curr. Treat. Options in Oncol. 23, 1522–1534 (2022). https://doi.org/10.1007/s11864-022-01021-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11864-022-01021-8