Opinion statement
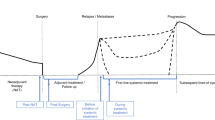
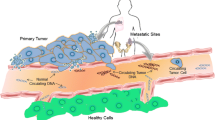
Liquid biopsy using blood components to assess circulating tumor DNA (ctDNA) is rapidly becoming a new standard-of-care technology in many tumor types, including breast cancer, due to the potential to provide predictive and prognostic information. The minimally invasive and repeatable nature of plasma based mutational testing is appealing for patients and facilitates enhanced disease monitoring. It is important for the clinician to understand the benefits and limitations of this emerging assay and the potential applications in breast cancer. Multiple technologies have been employed to assess breast cancer ctDNA with high sensitivity and specificity leading to assays that have been useful in research trials and are entering widespread clinical application. ctDNA analysis of breast cancer is of clinical utility today in selecting targeted therapy for advanced breast cancer, most notably by assessing PIK3CA mutations in hormone receptor–positive, HER2-negative disease. It will be employed in the near future in a variety of clinical settings including early detection of primary breast cancer, minimal residual disease after initial therapy, and use in advanced breast cancer for prognosis, early identification of non-response, and monitoring genomic markers of resistance.
Similar content being viewed by others
References and recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4(6):650–61. https://doi.org/10.1158/2159-8290.CD-13-1014.
Alimirzaie S, Bagherzadeh M, Akbari MR. Liquid biopsy in breast cancer: a comprehensive review. Clin Genet. 2019;95(6):643–60. https://doi.org/10.1111/cge.13514.
Therascreen PIK3CA RGQ PCR Kit Instructions for Use (Handbook) In: Qiagen, editor. 2019.
Juric D, Janku F, Rodon J, Burris HA, Mayer IA, Schuler M, et al. Alpelisib plus fulvestrant in PIK3CA-altered and PIK3CA-wild-type estrogen receptor-positive advanced breast cancer: a phase 1b clinical trial. JAMA Oncol. 2019;5(2):e184475. https://doi.org/10.1001/jamaoncol.2018.4475.
•• Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380(20):1929–40. https://doi.org/10.1056/NEJMoa1813904. Phase 3 trial demonstrating value of ctDNA in selecting targeted therapy based on PIK3CA mutation status.
O'Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9(1):896. https://doi.org/10.1038/s41467-018-03215-x.
Rothe F, Silva MJ, Venet D, Campbell C, Bradburry I, Rouas G, et al. Circulating tumor DNA in HER2-amplified breast cancer: a translational research substudy of the NeoALTTO phase III trial. Clin Cancer Res. 2019;25(12):3581–8. https://doi.org/10.1158/1078-0432.CCR-18-2521.
Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, et al. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells. Clin Cancer Res. 2017;23(20):6086–93. https://doi.org/10.1158/1078-0432.CCR-17-1173.
Fribbens C, Garcia Murillas I, Beaney M, Hrebien S, O'Leary B, Kilburn L, et al. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann Oncol. 2018;29(1):145–53. https://doi.org/10.1093/annonc/mdx483.
Bidard FC, Callens C, Pistilli B, Dalenc F, de La Motte RT, Sabatier R, et al. Emergence of ESR1 mutation in cell-free DNA during first line aromatase inhibitor and palbociclib: an exploratory analysis of the PADA-1 trial. Ann Oncol. 2019;30:v105–v6. https://doi.org/10.1093/annonc/mdz242.002.
Carausu M, Bidard FC, Callens C, Melaabi S, Jeannot E, Pierga JY, et al. ESR1 mutations: a new biomarker in breast cancer. Expert Rev Mol Diagn. 2019;19(7):599–611. https://doi.org/10.1080/14737159.2019.1631799.
Condorelli R, Spring L, O'Shaughnessy J, Lacroix L, Bailleux C, Scott V, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol. 2018;29(3):640–5. https://doi.org/10.1093/annonc/mdx784.
Stover DG, Parsons HA, Ha G, Freeman SS, Barry WT, Guo H, et al. Association of cell-free DNA tumor fraction and somatic copy number alterations with survival in metastatic triple-negative breast cancer. J Clin Oncol. 2018;36(6):543–53. https://doi.org/10.1200/JCO.2017.76.0033.
Ben Lassoued A, Nivaggioni V, Gabert J. Minimal residual disease testing in hematologic malignancies and solid cancer. Expert Rev Mol Diagn. 2014;14(6):699–712. https://doi.org/10.1586/14737159.2014.927311.
Czyz A, Nagler A. The role of measurable residual disease (MRD) in hematopoietic stem cell transplantation for hematological malignancies focusing on acute leukemia. Int J Mol Sci. 2019;20(21). https://doi.org/10.3390/ijms20215362.
McDonald BR, Contente-Cuomo T, Sammut SJ, Odenheimer-Bergman A, Ernst B, Perdigones N, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med. 2019;11(504). https://doi.org/10.1126/scitranslmed.aax7392.
• Garcia-Murillas I, Chopra N, Comino-Mendez I, Beaney M, Tovey H, Cutts RJ, et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. 2019. https://doi.org/10.1001/jamaoncol.2019.1838. Prospective determination of ctDNA predicting clinical relapse after primary therapy.
Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, et al. Personalized detection of circulating tumor DNA antedates breast Cancer metastatic recurrence. Clin Cancer Res. 2019;25(14):4255–63. https://doi.org/10.1158/1078-0432.CCR-18-3663.
Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. https://doi.org/10.1126/scitranslmed.aab0021.
Sparano JA, Henry NL. Surveillance after treatment of localized breast cancer: time for reappraisal? J Natl Cancer Inst. 2019;111(4):339–41. https://doi.org/10.1093/jnci/djy153.
Madic J, Kiialainen A, Bidard F-C, Birzele F, Ramey G, Leroy Q, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer. 2015;136(9):2158–65. https://doi.org/10.1002/ijc.29265.
Rodriguez BJ, Cordoba GD, Aranda AG, Alvarez M, Vicioso L, Perez CL, et al. Detection of TP53 and PIK3CA mutations in circulating tumor DNA using next-generation sequencing in the screening process for early breast cancer diagnosis. J Clin Med. 2019;8(8). https://doi.org/10.3390/jcm8081183.
• Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–30. https://doi.org/10.1126/science.aar3247. Promising development of a test to detect multiple cancers, including breast cancer, with blood.
Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403). https://doi.org/10.1126/scitranslmed.aan2415.
Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54(2):414–23. https://doi.org/10.1373/clinchem.2007.095992.
Shan M, Yin H, Li J, Li X, Wang D, Su Y, et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget. 2016;7(14):18485–94. https://doi.org/10.18632/oncotarget.7608.
Bacolod MD, Huang J, Giardina SF, Feinberg PB, Mirza AH, Swistel A, et al. Prediction of blood-based biomarkers and subsequent design of bisulfite PCR-LDR-qPCR assay for breast cancer detection. BMC Cancer. 2020;20(1):85. https://doi.org/10.1186/s12885-020-6574-4.
Jung A, Kirchner T. Liquid biopsy in tumor genetic diagnosis. Dtsch Arztebl Int. 2018;115(10):169–74. https://doi.org/10.3238/arztebl.2018.0169.
Palmirotta R, Lovero D, Cafforio P, Felici C, Mannavola F, Pelle E, et al. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:1758835918794630. https://doi.org/10.1177/1758835918794630.
Chin RI, Chen K, Usmani A, Chua C, Harris PK, Binkley MS, et al. Detection of solid tumor molecular residual disease (MRD) using circulating tumor DNA (ctDNA). Mol Diagn Ther. 2019;23(3):311–31. https://doi.org/10.1007/s40291-019-00390-5.
Jaiswal S, Ebert BL. MDS is a stem cell disorder after all. Cancer Cell. 2014;25(6):713–4. https://doi.org/10.1016/j.ccr.2014.06.001.
Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. https://doi.org/10.1182/blood-2015-03-631747.
Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484. https://doi.org/10.1038/ncomms12484.
Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130(6):742–52. https://doi.org/10.1182/blood-2017-02-769869.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Emily Miller declares that she has no conflict of interest. Lee Schwartzberg has received compensation from Amgen, Pfizer, Helsinn, Genentech/Roche, Genomic Health, Bristol-Myers Squibb, Myriad, AstraZeneca, and Bayer for service as a consultant.
Human and Animal Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Breast Cancer
Rights and permissions
About this article
Cite this article
Miller, E., Schwartzberg, L. Precision Medicine for Breast Cancer Utilizing Circulating Tumor DNA: It Is in the Blood. Curr. Treat. Options in Oncol. 21, 89 (2020). https://doi.org/10.1007/s11864-020-00783-3
Published:
DOI: https://doi.org/10.1007/s11864-020-00783-3