Abstract
Gramine is an indole alkaloid found in certain grass species. Results of previous studies have diverged as to whether this compound might cause resistance to aphids or not. In a breeding program aiming to introduce resistance to the cereal pest Rhopalosiphum padi L. in barley, a cultivar has been crossed with the progenitor of cultivated barley, Hordeum vulgare ssp. spontaneum, and an F1-derived population of doubled haploid (DH) lines was screened both for seedling gramine concentration and resistance to R. padi. The resistance was measured as individual aphid growth in the laboratory. The present study aims to determine if there is a genetic relationship between aphid growth retardation and gramine concentration. To do so, the lines were genotyped with a 384 SNP oligonucleotide pool assay and QTL analyses were performed for both traits. A previously identified aphid resistance locus on the distal part of chromosome 2HS was thereby confirmed, with resistance inherited from H. v. ssp. spontaneum, whereas one or more QTL for gramine concentration were potentially indicated on chromosome 3H, thus corroborating that the two traits are not linked genetically.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Small grain cereals in temperate regions on all continents are hosts of the bird cherry—oat aphid, Rhopalosiphum padi L. (Blackman and Eastop 2007). This pest causes direct damage via feeding on phloem sap but more seriously the aphid is also an important vector for barley yellow dwarf and cereal yellow dwarf viruses (Jarosova et al. 2016). Thus, it is important to be able to control R. padi and at present this is primarily achieved with insecticides. However, control via host plant resistance is a desirable alternative or complement, but this is difficult to breed for since R. padi feeding does not cause conspicuous leaf symptoms unlike Russian wheat aphid (RWA; Diuraphis noxia Kurdjumov) or greenbug (GB; Schizaphis graminum Rondani). Resistant barley cultivars have been bred to control these two other aphid species, with resistant plants being selected based on typical leaf symptoms such as chlorosis (RWA and GB) and leaf rolling (RWA), and plant death (RWA and GB) (Mornhinweg et al. 2012, 2017). Resistance to R. padi is manifested as reduced aphid growth, reduced host acceptance or increased plant tolerance. Those quantitative traits are cumbersome to measure and consequently there are no documented examples of cultivars bred for resistance to R. padi. However, there are identified gene sources for resistance to R. padi in cereals such as barley (Porter et al. 1999), including accessions of the progenitor of cultivated barley, Hordeum vulgare ssp. spontaneum (Weibull 1994a; Åhman et al. 2000; Ninkovic and Åhman 2009).
If the plant traits causing the resistance are known, then the selection of parents in a breeding program could be based on those, rather than on aphid performance in time-consuming aphid tests. There have been studies trying to elucidate which morphological (e.g. Tsumuki et al. 1989; Moharramipour et al. 1997a) and chemical (e.g. Weibull 1994b) plant characteristics are responsible for resistance to R. padi in barley. Several studies reviewed by Corcuera (1993) have found correlations between concentration of the indole alkaloid gramine in diet or plants and aphid responses such as reduced ingestion, longevity, fecundity and survival rate; with R. padi included among the aphid species studied. However, there are other studies where no relationship between gramine concentration and aphid responses has been found (Hautala and Holopainen 1995; Moharramipour et al. 1997a, b; Forslund et al. 1998). This was also the case when Åhman et al. (2000) attempted to use high gramine concentration as a chemical marker for R. padi resistance in a breeding program, finding no correlation between gramine concentrations and levels of aphid resistance measured as reduced aphid growth or reduced host plant acceptance. In that study, however, one accession was found, among several high-gramine H. v. ssp. spontaneum accessions, which did exhibit reduced aphid weight to approximately half of that on barley cultivars. This gene source accession denoted H. spont. 5 had been crossed with the cultivar Lina, one of the few Swedish two-rowed barley cultivars with moderate concentrations of gramine (Åhman et al. 2000). A doubled haploid (DH) line population developed from the F1 of this cross is the foundation of a long-term breeding program to introduce aphid resistance into cultivated barley using reduced nymphal weight gain when selecting for resistance (Åhman and Bengtsson 2019). One reason for using low individual aphid weight after feeding for a certain time period on a test plant as a measure of plant resistance is that small females carry few embryos and are, therefore, expected to have low birth rate (Dewar 1977). A population growth simulation study has shown that a 20% reduction in R. padi birth rate during seedling to beginning of ear emergence is expected to reduce the aphid population size by 40% when the population is at its peak. Alternatively, if the reduced aphid weight is due to longer development time, a 20% prolongation of time period until adulthood will reduce the peak population size by more than 50% (Wiktelius and Pettersson 1985). To validate the phenotyping method used for assessing barley resistance to R. padi in the laboratory, a field experiment was performed including three DH lines produced after three back-crosses (BC) to Lina, three DH lines from “BC” to other cultivars, and the four susceptible cultivars used as female parents in BC. The field results confirmed that the resistance selection method based on individual aphid growth was useful in that R. padi density close to population peak on the 10 barley genotypes correlated with laboratory aphid weight data (Åhman and Bengtsson 2019). Furthermore, a recent study of the effects of the resistance in H. spont. 5 compared to a susceptible cultivar, showed reduced nymph mass gain and intrinsic rate of population increase due to phloem- and mesophyll factors that limit phloem sap ingestion in R. padi (Leybourne et al. 2019).
The present study utilizes the F1DH population from the cross between H. spont. 5 and Lina to dissect out the genetic control of both aphid growth retardation and gramine concentration and determine if there is a relationship between them or not.
Materials and methods
Plant material
The aphid resistance source was a Hordeum vulgare ssp. spontaneum accession (denoted as H. spont. 5) from Canada Park in Israel (Åhman et al. 2000). Anther culture technique was used for DH production from the F1 and the DH lines were propagated from DH mother plants in the greenhouse. These were kept in perforated plastic bags during flowering and seed development to hinder cross-pollination. Eighty-eight DH lines were tested for aphid resistance and gramine concentration at seedling stage.
Aphid tests
Colonies of R. padi were reared on oats in cages in a greenhouse, with lighting regime and temperature simulating summer conditions. Initially, the rearing was started from spring migrants on the winter host, Prunus padus L., ensuring that aphids were free of yellow dwarf viruses (BYDV/CYDV).
Seeds for test plants were soaked with 0.75% H2O2 in water on filter paper in Petri dishes and kept for 3 days in a refrigerator. After 2 days at room temperature in the laboratory, seedlings were transplanted to slow-release-fertilized (1 dl of 15-4.8-10.8 NPK Osmocote Plus and 15 ml Micromax minerals per 50 l) Hammenhög’s potting soil in 10 cm diameter plastic pots. One potted plant per DH line was placed in each of four white plastic trays (41 × 62 × 11 cm) in a random order, each tray representing one replicate. The trays were placed in a Conviron growth cabinet E15, CMP 3244; temperature 22 °C, 16 h light at 220 µmol photons/m2/s at plant level and minimum 80% RH. After another 2 days, cylindrical Perspex tubes (2 cm diameter, 5 cm high) were slipped over each seedling allowing the plant to grow through. The tests started 1 week after transplanting by adding 5 newborn nymphs to each tube cage which was then sealed with cotton wool at the top. The nymphs were offspring born on oats by alate females collected on the walls of the rearing cages the day before. After 4 days, the nymphs were weighed singly on a Mettler M3 microbalance. Due to space limitation in the Conviron cabinet and the time needed for weighing the aphids, all the 88 DH lines could not be tested simultaneously. Each of the five tests included between 11 and 23 test DH lines together with the parental lines cultivar Lina and H. spont. 5. To compare between tests, mean weight of up to five recovered aphids was divided by the mean weight for the recovered aphids on the control Lina in each replicate, and the mean quota for each line was entered in the QTL analysis.
Gramine analysis
Plants for gramine analysis were grown in the same Conviron cabinet and under the same conditions as for aphid tests except that temperature was higher, 25 °C, to accentuate gramine-level differences (Hanson et al. 1983). Due to space limitations in the growth cabinet, cultures were separated in five rounds, described in more detail by Åhman et al. (2000). In four of the culture rounds, seedlings of all five test plants in each of two replicates were cut just above the apex of the coleoptile and stored in a freezer (− 18 °C), from there taken to a − 35 °C freezer for 1 day and thereafter freeze-dried for three days and then milled. Five mg of this flour was mixed with 5 ml of purified water (Millipore) and heated in a water bath (70 °C) for 15 min. during which time the samples were shaken three times. 200 µl of the supernatant obtained after centrifuging at 3000×g for 20 min. was mixed with 800 µl of water. From the fifth culture round, freeze-dried plants were analyzed singly by extracting gramine in 20 ml of water while shaken in steel tubes with two steel bullets at room temperature for 1 h. 750 µl of the supernatant was mixed with water 1:1. Each of the two replicates was in this test round based on means of 4 plants. The reason for using water for gramine extractions rather than other less user-friendly and more expensive solvents was that the method was intended for routine screenings of thousands of lines, if a positive relationship between gramine concentration and resistance to R. padi was found and gramine-based selections were to be used in practical breeding. The single-plant extraction method would make use of fewer plants for phenotyping.
HPLC analysis was performed on a Milton Roy (LDC) chromatograph equipped with a C18, Guard Pak (Waters Nova-Pac) pre-column and a 150 mm × 3.9 mm, 4 µm grain size C18 column (Waters Nova-Pac). Twenty µl per sample were autoinjected. Eluents were: (A) 10% acetonitrile and 0.1% triethylamine in water with pH adjusted to 5.5 with acetic acid and (B) 50% acetonitrile in water. Gramine was eluted with a mixture of 15% A and 85% B at a flow rate of 1.5 ml/min. The eluate was analyzed at 218 nm with a Milton Roy UV SM 4000 detector. The retention time for gramine was ca. 3.1 min. The response factor and linearity were determined with gramine from Sigma at a concentration of 4, 8 and 12 µg/ml.
DNA extraction, SNP genotyping and analysis
For genotyping the population, genomic DNA was extracted from each plant of 88 F1DH lines using ~ 100 mg of leaf tissue collected from greenhouse-grown plants using the Qiagen DNAeasy Plant Maxi Kit (Qiagen, Valencia, CA, USA). A standard in-house 384 oligonucleotide pool assay (OPA) was used that was based on polymorphisms identified using Barley OPA 1 and Barley OPA 2 (Close et al. 2009) with the single-nucleotide polymorphisms (SNPs) represented on the OPA selected to maximize polymorphism and provide good genome coverage (Muñoz-Amatriaín et al. 2011). Genotyping was performed using an Illumina BeadXpress at the James Hutton Institute (Invergowrie, Scotland, UK) and the results analyzed using the Illumina Genome Studio software (Genotyping Analysis Module v1.8.4). The graphical genotypes produced were used to double check the allele calls manually to verify all recombination breakpoints.
Linkage mapping JoinMap 4 (Kyazma B.V.) was used for de novo construction of linkage maps (Van Ooijen 2006) using the 223 markers polymorphic in the population. A minimum logarithm of the odds ratio (LOD) score of 4 was used in the formation of linkage groups. The maximum likelihood mapping algorithm was used to determine marker order in each linkage group. Recombination frequencies were converted to centiMorgans (cM) using Kosambi’s mapping function as implemented in JoinMap 4 for visualization.
QTL interval mapping was performed using MapQTL 5 (https://www.kyazma.nl/index.php/MapQTL/; accessed on 6 March 2019) with a subset of the SNP genotypic data to provide full-genome coverage with minimal redundancy. Permutation tests with 10,000 permutations were used to establish the LOD thresholds. Restricted multiple QTL mapping (rMQM mapping) was used to search for further QTLs after taking into account the most significant ones.
Results
The cross between the cultivar Lina and H. spont. 5 exhibited polymorphism at 223 out of the 384 SNP loci on the OPA assay, with 24, 32, 31, 29, 47, 33 and 27 SNP distributed on the chromosomes 1H, 2H, 3H, 4H, 5H, 6H and 7H, respectively (Fig. 1). The coverage of the genome was near complete with the mapped polymorphic SNPs representing 94.4% of consensus genetic map (Muñoz-Amatriaín et al. 2011).
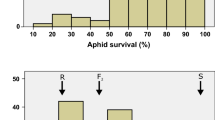
Mean aphid weight on the 88 DH lines varied between 64 and 176% of that on cultivar Lina (Fig. 2). QTL analysis detected a significant QTL on the very distal part of chromosome 2HS associated with the SNP 11_11059 at 0 cM with a LOD score of 4.7 (Fig. 3) explaining 24.9% of the variance in aphid weight relative to that on the control Lina, and with the H. spont. 5 allele associated with the lower aphid weight (Fig. 3).
Mean gramine concentration in seedlings of the DH lines varied between 13 and 24 mg/g dry weight (Fig. 4). Parent Lina’s mean values varied between 9 and 12 and those of parent H. spont. 5 between 15 and 23 in the five test rounds. The lowest parent values were from the single-plant-extraction test, indicating that this extraction procedure was less efficient. However, there was variation between the eight DH lines subjected to this extraction method too, with means ranging from 13 to 20 mg/g dry weight (Fig. 4). QTL analysis of gramine concentrations in the DH population detected multiple peaks between 100 and 140 cM on chromosome 3H at maximum LOD scores of 2.3, below the LOD significance level at 2.8 determined by permutation tests (Fig. 5). The region associated with this non-significant QTL showed higher gramine values to be associated with the H. spont. 5 alleles.
Distribution of the 88 F1DH lines in the Lina × H. spont. 5 population based on seedling mean concentration of gramine. Turquoise = line data from two replicates, with five plants in each extracted together; yellow = data from eight of the lines, represented by two replicates with four singly extracted plants in each
Chart of the LOD score obtained from interval mapping of gramine concentration in seedlings (mg/g dry matter) on chromosome 3H in the Lina x H. spont. 5 F1DH population. SNP designations on the x-axis and LOD values on the y-axis. Dotted line indicates the threshold for significance obtained by permutation test
Discussion
Gramine occurs in certain species of Hordeum but also in a few other grass species such as Phalaris arundinacea L. and Arundo donax L. (Kokubo et al. 2017). Many barley cultivars contain no or just trace amounts of gramine in seedlings whereas others contain moderate levels, as does the cultivar Lina (Hanson et al. 1981; Forslund et al. 1998; Åhman et al. 2000; Larsson et al. 2006). Concentrations in the wild progenitor of cultivated barley, H. v. ssp. spontaneum, also vary. The range was 5–23 mg/g dry matter in a set of 157 different accessions (Åhman et al. 2000); however, none was found with no or just trace levels of gramine. Relatively high levels were also found in the four accessions of H. v. ssp. spontaneum studied by Hanson et al. (1981) as well as those studied by Moharramipour et al. (1999).
Gramine biosynthesis starts from tryptophan, with 3-aminomethylindole (AMI) and N-methyl-3-aminomethylindole (MAMI) further down in the pathway (Gross et al. 1974). Based on comparisons between two cultivars, Arimar and Proctor; with and without gramine accumulation, respectively; Hanson et al. (1983) proposed that gramine synthesis may be dependent on a single gene of importance for converting tryptophan to a precursor of AMI. In a follow-up study, Leland et al. (1985) included a third cultivar, Morex, also without gramine. This cultivar is in addition deficient in a specific N-methyltransferase (NMT), essential for the conversion of AMI to MAMI and MAMI to gramine (Larsson et al. 2006), whereas Proctor is not. F1 offspring from crosses between the two gramine-free cultivars and Arimar had mid-parent gramine concentrations. A two-gene model was tested in the F2-generation from the cross between Morex and Arimar, but the offspring segregated in a 1:3 ratio (no gramine:full gramine synthesis) suggesting that the two genes are tightly linked (Leland et al. 1985). The bimodal 1:1 distribution of gramine data in the Steptoe/Morex DH population studied by Yoshida et al. (1997) supports this conclusion.
Moharramipour et al. (1999) studied other crosses between low-level and high-level gramine parents and found evidence of both a mono- and a di-gene model with additional minor genes controlling the gramine concentrations in gramine-containing plants at adult stage. They found partial dominance for lower levels of gramine in their studies of adult plants. On the contrary, crosses with the moderate-level Lina and the high-gramine H. spont. 5, as well as with several other H. v. ssp. spontaneum accessions, resulted in bias towards higher levels of gramine in seedlings (Åhman et al. 2000), suggesting that parents contributed partly different alleles for high-gramine concentration in seedlings. Both of the parents are known to express the NMT protein (Larsson et al. 2006). The present QTL analysis of the Lina × H. spont. 5 F1 DH population, indicates that there may be genes on chromosome 3H controlling gramine concentrations in seedlings.
The present study also confirms a position of a locus for R. padi resistance distally on the short arm of barley chromosome 2H indicated in earlier studies using the same Lina × H. spont. 5 barley population, but analyzed with other molecular marker systems (Louise O’Donoghue, unpublished; Åhman and Bengtsson 2019). The allele for aphid resistance at this QTL on 2HS was derived from the wild H. spont. 5 parent and we found no support for a genetic linkage between this aphid resistance and QTL for gramine concentration since loci controlling the phytochemical trait were potentially indicated on chromosome 3H.
In the effort to introgress the aphid resistance from the wild barley source H. spont. 5 into cultivated barley, DH lines with a minor resistance effect have been found despite that they do not contain the 2HS resistance QTL (Åhman and Bengtsson 2019). When a BC2F1 DH population originating from a BC1F1 DH line lacking the marker associated with the H. spont. 5 resistance QTL on 2HS was analyzed, a QTL for R. padi resistance was found on 3H in a pericentromeric position at 76 cM, with the resistance allele contributed by Lina (Cheung et al. 2010). However, this QTL was not evident in the present study using the F1 DH population from the same pedigree, potentially due to the limited size of the mapping population.
Interestingly, the present study does indicate potential QTL for gramine concentration on 3H albeit below the LOD threshold for significance. However, this region is on the long arm of this chromosome and appears to derive from the H. spont. 5 parent so there is no genetic support for a relationship between high-gramine concentration and seedling resistance to the aphid R. padi in this barley breeding material. Although the QTL for gramine concentration did not reach significance threshold, positive alleles at gramine-controlling loci were expected from both parents given earlier work comparing the phenotype across a range of barley germplasm (Åhman et al. 2000).
Similar to our results, there was no support for a genetic relationship between gramine concentration (Yoshida et al. 1997) and aphid resistance (Moharramipour et al. 1997b) in a DH population derived from a cross between cultivars Steptoe and Morex. In this population, a single major effect locus for gramine concentration was found at the centromere region of 1H (formerly denominated as chromosome 5) that corresponds to the position of the NMT gene missing in Morex (Lee et al. 1997; Larsson et al. 2006). Loci controlling aphid resistance were also found in this population (Moharramipour et al. 1997b); one in the centromeric region of chromosome 2H as well as one in the long arm of 1H, distant from the gramine QTL on this chromosome. The aphid data were collected in field trials from 1994, 1995 and 1996 and the aphid species composition varied. When R. padi dominated in 1996, no QTL for aphid resistance were detected, indicating that the QTL on 1H and 2H potentially harbored genes acting in resistance to one or more of the other aphid species, which were predominantly Rhopalosiphum maidis Fitch, but also Sitobion akebiae Shinji and GB.
Using a DH population derived from another cross, Harrington/TR306, two aphid resistance QTL were mapped to the distal end of the short arms of chromosomes 1H and 7H (formerly denominated as chromosome 1; Moharramipour et al. 1997c). The aphid data were collected in field trials from 1994 and 1995 when R. maidis predominated but R. padi was also present. The region of this QTL on 1HS contains the aphid induced protease inhibitor CI2c (Wei et al. 2002) that has been the subject of over-expression studies in barley (Losvik et al. 2018), since it was one of four candidate genes for R. padi resistance from H. spont. 5 suggested by Delp et al. (2009). Interestingly, the behaviour and performance of R. padi were not affected by changes in the expression of CI2c, whereas the behaviour of another aphid species (Myzus persicae Sulzer) was altered (Losvik et al. 2018).
QTL analyses and genome-wide association mapping of RWA resistance in barley have revealed several responsible loci; among them loci on chromosome 2H and 3H (Nieto-Lopez and Blake 1994; Mittal et al. 2008, 2009; Tocho et al. 2012, 2013; Dahleen et al. 2015). The RWA resistance loci on 2H is at 27 and 39 cM (Mittal et al. 2008; Mittal et al. 2009, respectively), 18 and 20 cM (Dahleen et al. 2015), 70–96 cM (Tocho et al. 2012) and at 70–96 cM for GB resistance as well (Tocho et al. 2013) and thus not at the same locus as the present QTL for R. padi resistance. The RWA resistance locus on 3H is at around 147 and 108 cM (Mittal et al. 2008 , 2009, respectively) and 136 cM (Dahleen et al. 2015), potentially within the region of the gramine QTL in our study. Resistance to GB was also located in that region of 3H, at ca. 140 cM, but the suggested candidate resistance gene is a nucleotide-binding site—leucine-rich repeat gene (NBS-LRR; Azhaguvel et al. 2014).
Several NBS-LRR sequences (R-genes) are located in the 0–6 cM QTL for R padi resistance at 2HS (Åhman and Bengtsson 2019). This locus also harbours other candidate genes for resistance such as genes involved in ethylene signalling, a cysteine-rich receptor-like protein kinase gene (R-gene) and a laccase gene potentially involved in lignification (Åhman and Bengtsson 2019). Ongoing R-gene capturing experiments in breeding lines with and without the 2HS resistance haplotype from H. spont. 5 will hopefully help reveal whether it is likely that such genes are involved in causing the 2HS-related resistance to R. padi. Meanwhile, the substantially denser SNP array used in the study by Åhman and Bengtsson (2019) than in the present study has revealed several new SNP markers distally at chromosome 2HS, markers that are presently being used in practical breeding for resistance to R. padi to partly substitute for the cumbersome aphid resistance phenotyping method based on reduced nymphal weight.
References
Åhman I, Bengtsson T (2019) Introgression of resistance to Rhopalosiphum padi L. from wild barley into cultivated barley facilitated by doubled haploid and molecular marker techniques. Theor Appl Genet. https://doi.org/10.1007/s00122-019-03287-3
Åhman I, Tuvesson S, Johansson M (2000) Does indole alkaloid gramine confer resistance in barley to the aphid Rhopalosiphum padi? J Chem Ecol 26:233–255
Azhaguvel P, Mornhinweg D, Vidya-Saraswathi D, Rudd JC, Chekhovskiy K, Saha M, Close TJ, Dahleen LS, Weng Y (2014) Molecular mapping of greenbug (Schizaphis graminum) resistance gene Rsg1 in barley. Plant Breed 133:227–233
Blackman RL, Eastop VF (2007) Taxonomic Issues. In: Van Emden HF, Harrington J (eds) Aphids as Crop Pests. CAB International, Cromwell Press, Trowbridge, pp 1–29
Cheung WY, Di Giorgio L, Åhman I (2010) Mapping resistance to the bird cherry-oat aphid (Rhopalosiphum padi) in barley. Plant Breed 129:637–646. https://doi.org/10.1111/j.1439-0523.2010.01771.x
Close TJ, Bhat PR, Lonardi S et al (2009) Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10:582. https://doi.org/10.1186/1471-2164-10-582
Corcuera LJ (1993) Biochemical basis for the resistance of barley to aphids. Phytochemistry 33:741–747
Dahleen LS, Bregitzer P, Mornhinweg D, Esvelt Klos K (2015) Genetic diversity for Russian wheat aphid resistance as determined by genome-wide association mapping and inheritance in progeny. Crop Sci 55:1925–1933. https://doi.org/10.2135/cropsci2014.09.0634
Delp G, Gradin T, Åhman I, Jonsson LMV (2009) Microarray analysis of the interaction between the aphid Rhopalosiphum padi and host plants reveals both differences and similarities between susceptible and partially resistant barley lines. Mol Genet Genomics 281:233–248. https://doi.org/10.1007/s00438-008-0409-3
Dewar AM (1977) Assessment of methods for testing varietal resistance to aphids in cereals. Ann Appl Biol 87:183–190. https://doi.org/10.1111/j.1744-7348.1977.tb01874.x
Forslund K, Pettersson J, Ahmed E, Jonsson L (1998) Settling behaviour of Rhopalosiphum padi (L.) in relation to cyanogenic glycosides and gramine contents in barley. Acta Agric Scand Sect B Soil Plant Sci 48:107–112
Gross DH, Lehmann H-R, Shutte PM (1974) Zur biosynthese des gramins. Biochem Physiol Pflanzen 166:281–287
Hanson AD, Traynor PL, Ditz KM, Reicosky DA (1981) Gramine in barley forage—effects of genotype and environment. Crop Sci 21:726–730
Hanson AD, Ditz KM, Singletary GW, Leland TJ (1983) Gramine accumulation in leaves of barley grown under high-temperature stress. Plant Physiol 71:896–904
Hautala E-L, Holopainen JK (1995) Gramine and free amino acids as indicators of fluoride-induced stress in barley and its consequences to insect herbivory. Ecotoxicol Environ Saf 31:238–245
Jarosova J, Beoni E, Kumar Kundu J (2016) Barley yellow dwarf virus resistance in cereals: approaches, strategies and prospects. Field Crops Res 198:200–214. https://doi.org/10.1016/j.fcr.2016.08.030
Kokubo Y, Nishizaka M, Ube N, Yabuta Y, Tebayashi S, Ueno K, Taketa S, Ishihara A (2017) Distribution of the tryptophan pathway-derived defensive secondary metabolites gramine and benzoxazinones in Poaceae. Biosci Biotechnol Biochem 81:431–440
Larsson KAE, Zetterlund I, Delp G, Jonsson LMV (2006) N-methyltransferase involved in gramine biosynthesis in barley: cloning and characterisation. Phytochemistry 67:2002–2008
Lee JE, Kleinhofs A, Graner A, Wegener S, Parthier B, Löbler M (1997) Genomic sequence and mapping of a methyljasmonate-induced O-methyltransferase from barley (Hordeum vulgare L.). DNA Seq 7:357–363. https://doi.org/10.3109/10425179709034056
Leland TJ, Grumet R, Hanson AD (1985) Biochemical, immunological and genetic characterization of natural gramine-free variants of Hordeum vulgare L. Plant Sci 42:77–82
Leybourne DJ, Valentine TA, Robertson JAH, Pérez-Fernández E, Main AM, Karley AJ, Bos JIB (2019) Defence gene expression and phloem quality contribute to mesophyll and phloem resistance to aphids in wild barley. J Exp Bot 70:4011–4026. https://doi.org/10.1093/jxb/erz163
Losvik A, Beste L, Stephens J, Jonsson L (2018) Overexpression of the aphid-induced serine protease inhibitor CI2c gene in barley affects the generalist green peach aphid, not the specialist bird cherry-oat aphid. PLoS ONE 13(3):e0193816. https://doi.org/10.1371/journal.pone.0193816
Mittal S, Dahleen LS, Mornhinweg D (2008) Locations of quantitative trait loci conferring Russian wheat aphid resistance in barley germplasm STARS-9301B. Crop Sci 48:1452–1458. https://doi.org/10.2135/cropsci2007.11.0651
Mittal S, Dahleen LS, Mornhinweg D (2009) Barley germplasm STARS-9577B lacks a Russian wheat aphid resistance allele at a quantitative trait locus present in STARS-9301B. Crop Sci 49:1999–2004
Moharramipour S, Tsumuki H, Sato K, Murata S, Kanehisa K (1997a) Effects of color, epicuticular wax amount and gramine content in barley hybrids on cereal aphid populations. Appl Entomol Zool 32:1–8
Moharramipour S, Yoshida H, Sato K, Takeda K, Iida T, Tsumuki H (1997b) Mapping cereal aphid resistance in Steptoe/Morex doubled haploid population. Barley Genetics Newsletter 27, p 2
Moharramipour S, Tsumuki H, Sato K, Yoshida H (1997c) Mapping resistance to cereal aphids in barley. Theor Appl Genet 94:592–596
Moharramipour S, Takeda K, Sato K, Yoshida H, Tsumuki H (1999) Inheritance of gramine content in barley. Euphytica 106:181–185
Mornhinweg DW, Obert DE, Carver BF (2012) Registration of eight six-rowed feed barley germplasm lines resistant to both Russian wheat aphid and greenbug. J Plant Reg 6:186–189. https://doi.org/10.3198/jpr2011.09.0517crg
Mornhinweg DW, Hammon RW, Obert DE (2017) Registration of “Mesa” Russian wheat aphid-resistant winter feed barley. J Plant Reg 11:85–88. https://doi.org/10.3198/jpr2016.09.0050crc
Muñoz-Amatriaín M, Moscou MJ, Bhat PR, Svensson JT, Bartoš J, Suchánková P, Šimková H, Endo TR, Fenton RD, Lonardi S, Castillo AM, Chao S, Cistué L, Cuesta-Marcos A, Forrest KL, Hayden MJ, Hayes PM, Horsley RD, Makoto K, Moody D, Sato K, Vallés MP, Wulff BBH, Muehlbauer GJ, Doležel J, Close TJ (2011) An improved consensus linkage map of barley based on flow-sorted chromosomes and single nucleotide polymorphism markers. Plant Genome 4:238–239
Nieto-Lopez RM, Blake TK (1994) Russian wheat aphid resistance in barley: inheritance and linked molecular markers. Crop Sci 34:655–659
Ninkovic V, Åhman IM (2009) Aphid acceptance of Hordeum genotypes is affected by plant volatile exposure and is correlated with aphid growth. Euphytica 169:177–185. https://doi.org/10.1007/s10681-009-9918-3
Porter DR, Mornhinweg DW, Webster JA (1999) Insect resistance in barley germplasm. In: Clement SL, Quisenberry SS (eds) Global plant genetic resources for insect-resistant crops. CRC Press LLC, Boca Raton, pp 51–61
Tocho E, Ricci M, Tacaliti MS, Giménez DO, Acevedo A, Lohwasser U, Börner A, Castro AM (2012) Mapping resistance genes conferring tolerance to RWA (Diuraphis noxia) in barley (Hordeum vulgare). Euphytica 188:239–251. https://doi.org/10.1007/s10681-012-0710-4
Tocho E, Börner A, Lohwasser U, Castro AM (2013) Mapping and candidate gene identification of loci determining tolerance to greenbug (Schizaphis graminum, Rondani) in barley. Euphytica 191:173–182. https://doi.org/10.1007/s10681-012-0740-y
Tsumuki H, Kanehisa K, Kawada K (1989) Leaf surface was as a possible resistance factor of barley to cereal aphids. Appl Ent Zool 24:295–301
Van Ooijen (2006) JoinMap ® 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen, Netherlands
Wei F, Wing RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14:1903–1917. https://doi.org/10.1105/tpc.002238
Weibull J (1994a) Resistance to Rhopalosiphum padi L. (Homoptera: Aphididae) in Hordeum vulgare subsp. spontaneum and in hybrids with H. vulgare subsp. vulgare. Euphytica 78:97–101
Weibull J (1994b) Glutamic acid content of phloem sap is not a good predictor of plant resistance to Rhopalosiphum padi. Phytochemistry 35:601–602
Wiktelius S, Pettersson J (1985) Simulations of bird cherry-oat aphid population dynamics: a tool for developing strategies for breeding aphid-resistant plants. Agric Ecosyst Environ 14:159–170
Yoshida H, Iida T, Sato K, Moharramipour S, Tsumuki H (1997) Mapping a gene for gramine synthesis in barley. Barley Genetics Newsletter 27, p 3
Acknowledgements
Open access funding provided by Swedish University of Agricultural Sciences. The Hordeum vulgare ssp. spontaneum accession was supplied by Dr. Louis Lehmann a former researcher at the Swedish University of Agricultural Sciences. Yvonne Andersson helped with the aphid test and Maj Johansson with gramine analysis at Svalöf AB, in Svalöv, Sweden. Dr. Stine Tuvesson assisted with doubled haploid production. This work has been financed by The Swedish Farmer’s Association, The Swedish Foundation for Strategic Environmental Research (Mistra) through the PlantComMistra programme, The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning FORMAS, the plant breeding company Svalöf Weibull AB (presently Lantmännen Lantbruk ek för) and The Swedish University of Agricultural Sciences. The work at the James Hutton Institute was supported by the Scottish Government’s Rural and Environment Science and Analytical Services Division work programme Theme 2 WP2.1.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jarmo K. Holopainen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Macaulay, M., Ramsay, L. & Åhman, I. Quantitative trait locus for resistance to the aphid Rhopalosiphum padi L. in barley (Hordeum vulgare L.) is not linked with a genomic region for gramine concentration. Arthropod-Plant Interactions 14, 57–65 (2020). https://doi.org/10.1007/s11829-019-09727-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11829-019-09727-7