Abstract
Gold nanoparticles have numerous applications, many of which are notable in industries. The biosynthesis of gold nanoparticles offers an easy, effective, green, and eco-friendly approach. In organisms capable of synthesizing nanoparticles, enzymes and proteins are responsible for the structural and functional modifications that lead to their formation. These include ABC transporter, peptide-binding proteins, which are dependent on abiotic parameters. This study uses the purified ABC transporter, peptide-binding protein transformed from Thermus scotoductus SA-01 and expressed in mesophilic Escherichia coli BL21 and thermophilic Thermus thermophilus HB27 hosts for the biosynthesis of gold nanoparticles at different concentrations, temperatures, and pH values. Gold nanoparticle formation was evaluated with a range of gold (III) concentrations (0–10 mM), incubated at temperatures ranging from 30–85 ºC and pH levels from 3.6–9.0. Transmission electron microscopy (TEM), energy dispersive X-ray spectrometry (EDX), and UV–Vis absorption spectroscopy were used to characterise the formation of nanoparticles. In all of the protein reactions, UV–Vis absorbance peaks at approximately 520–560 nm confirmed the formation of gold nanoparticles. Optimum nanoparticle synthesis was observed at pH values ranging from 5.5 to 9.0, gold (III) solution (HAuCl4) concentrations from 0.5–2.0 mM, and a maximum temperature of 65ºC in the mesophilic host and 85ºC in the thermophilic host, indicating the significance of temperature in both hosts for the expression and bioactivity of the purified ABC transporter protein. However, the biogenic formation of gold nanoparticles using E. coli and T. thermophilus hosts was not monodispersed, suggesting a necessity for further development of the procedure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nanoparticles are fascinating ultrafine particles with distinct physicochemical properties that could be used for a variety of applications in engineering, electrical, medical, and biological fields for simultaneous diagnosis and drug delivery, electroanalysis, hydrogen gas sensing, and cancer diagnosis and treatment. Additionally, they can serve as emulsions in oil production, cheap stabilisers, CO2 foams, and reservoir sensing (Salata 2004; Yezhelyev et al. 2006; Campbell and Compton 2010; Lee et al. 2011; Ullah et al. 2017; Ko and Huh 2019). Although the colloidal solutions of the metallic nanoparticulate substances are ultrafine, with one dimension less than 100 nm, the molecules that make them up are very complex. They are made up of three layers, which contribute to their extraordinary mechanical strength, electrical properties, and thermal conductivity for better performance, structural durability, and reliability (Khan et al. 2019). Among the commonly used classes of metal nanoparticles include silver (Ag), gold (Au), and platinum (Pt). However, gold nanoparticles get greater attention because of their chemical inertness, non-cytotoxicity, less-efficient interactions, and ability to manipulate gold particulate size (Asnag et al. 2019). The distinct nanoscale properties of gold are utilized in numerous fields, including catalysis, nonlinear optical devices, biolabeling, colours and coatings, optical recording media, and the biomedical industry (Shnoudeh et al. 2019). However, gold is predominantly used for jewellery production worldwide, nanotechnology can expand its industrial applications.
The use of hazardous chemicals in the preparation of nanoparticles is not only challenging in terms of its synthesis protocol but also environmentally unfriendly and expensive (Shamaila et al. 2016). As a result, the biosynthesized formation of ultrafine particles using microorganisms is believed to be promising, sustainable, non-toxic, and economical. However, the utilization of microorganisms for large-scale nanoparticle production is still in its infancy. These microbes vary from simple prokaryotic bacteria to complex eukaryotic fungi and yeasts. In addition, biologically synthesized nanoparticles are typically more polydisperse when compared to nanomaterials produced using chemicals (Sinha et al. 2009). Many microorganisms can use reductase enzymes to convert metal salts into monodispersed metal nanoparticles, detoxify them, and use the metal as an energy source. This is due to the microbe's inherent ability to facilitate energy-dependent proton ion efflux through the cell membrane using ATPase (Li et al. 2011).
Microorganisms, including bacteria, have been extensively studied for nanoparticle synthesis with an array of biological protocols that have been reported using various bacterial biomasses, mostly mesophiles (Singh et al. 2016). Some of these techniques, however, have many downsides and limitations, particularly when employed for the commercial synthesis of nanoparticles at high temperatures. Several thermostable enzymes require cofactors, active chaperones, and specific posttranslational processing for correct protein folding, which may not be optimal at high temperatures in a mesophilic host like Escherichia coli (Bhatwa et al. 2021). Thermophiles such as Thermus scotoductus and Thermus thermophilus have been proposed as reliable sources of industrial thermostable enzymes in biosynthesized nanoparticle formation (Turner et al. 2007). Thermus species are tolerant to thermophilic environments with high natural transformation and genetic plasticity, as well as being amenable to genetic manipulation due to their inherent ability to be transformed with chromosomal or plasmid DNA (Carr et al. 2015). The rapid growth rates and abundance of enzymes produced have made this organism useful in various biotechnological applications, primarily in the thermostabilizing of enzymes. Several factors, including temperature, pH, exposure time, biomass, and substrate concentration are used to manipulate and optimize nanoparticle properties and formation. These parameters may thus influence the growth conditions of the organisms, their cellular activities, and their enzymatic processes (Korbekandi et al. 2009).
Van Marwijk (2009) and Opperman et al. (2008) revealed in their studies that T. scotoductus SA-01 could reduce heavy metals with the subsequent formation of nanoparticles. In this case, soluble gold (III) was reduced to insoluble elemental gold. This thermophilic bacterium was isolated by Kieft et al. (1999) from groundwater samples from Mponeng (a deep South African gold mine in the Witwatersrand Supergroup operated by AngloGold Ashanti) at a depth of 3.2 km with an ambient rock temperature of 60 ºC and it was later described by Balkwill et al. (2004) as T. scotoductus. Thermus scotoductus SA-01 has several membrane proteins as well as cytoplasmic proteins that participate in the gold reduction and nanoparticle formation processes (Erasmus et al. 2014). In order to achieve a thermostable microbial strain capable of biosynthesis of nanomaterials, a cloning vector from the plasmid of Thermus sp. is required (Lasa et al. 1992; De Grado et al. 1999). The genome of T. scotoductus SA-01 is, however, related to that of T. thermophilus HB27, which is a commonly used thermophile strain. T. thermophilus HB27 consists of 1,894,877 bp chromosomes and 232, 605 bp megaplasmid (Henne et al. 2004), whereas T. scotoductus SA-01 consists of 2,346,803 bp chromosomes and 8,383 bp plasmids with approximately 69% G + C content similarity to T. thermophilus (Gounder et al. 2011), making the strains suitable for transformation.
Studies have identified that ABC transporter peptide-binding genes are membrane proteins, consisting of eight subfamilies (A-H), depending on their structural arrangement and phylogenetic analyses (Dean et al. 2001; Popovic et al. 2010). They are composed of four core domains, namely, two nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs). The former contains sequence motifs needed for ATP hydrolysis, whereas the latter comprises 4–10 α-helices and serves as a substrate translocation pathway across the lipid bilayer (Cai and Gros 2003). The four domains are either encoded on a single gene, one NBD and one TMD connected on a gene, or all the domains are encoded by separate genes (Rea 2007). ABC transporter, peptide-binding protein, is a reductase-like protein that has been identified as capable of reducing uranium and thus could have been the principal protein in the Thermus species that aids in reducing gold, chromate, iron, and uranium (Balkwill et al. 2004; Opperman and Van Heerden 2007; Gounder et al. 2011; Cason et al. 2012). In this study, ABC transporter peptide-binding protein identified in T. scotoductus SA-01 (Erasmus et al. 2014), was transformed in E. coli BL21 (DE3) and T. thermophilus HB27 as mesophilic and thermophilic hosts, respectively for gold reduction and nanoparticle formation. Effects of varying gold (III) concentrations, temperatures, and pH values were assessed on the gold reduction and nanoparticles formation. This was followed by the characterization of the obtained nanoparticles using transmission electron microscopic and energy dispersive X-ray spectrometric analyses.
Materials and methods
Bacterial strains, growth conditions and genomic DNA isolation
In this study, chromosomal DNA was isolated and transformed from T. scotoductus SA-01 (ATCC 700910) strain using an adapted SDS-proteinase procedure. E. coli TOP10 (Invitrogen) was used as a host for cloning of the protein while T. thermophilus HB27 (ATCC BAA-163) and E. coli BL21 (DE3) were used as hosts for the expression of the ABC transporter peptide-binding protein. T. scotoductus SA-01 (American Type Culture Collection) grown in glycerol stock (maintained at -80 ºC), was cultured in Tryptone, Yeast Extract, Glucose (TYG) broth [18 g L−1 bacteriological agar (Biolab), 5 g L−1 tryptone (Biolab), 3 g L−1 yeast extract (Biolab), and 1 g L−1 glucose (Saarchem), pH 7.0], and incubated, under aerobic conditions for 24 h at 65 ºC to reach the mid-exponential growth phase. E. coli TOP10 and E. coli BL21 (mesophilic host) cells were grown aerobically in Luria–Bertani (LB) medium with a final pH of 7.0 at 37 ºC. The LB medium was supplemented with 100 µg mL−1 ampicillin or 30 µg mL−1 kanamycin as required (Sambrook et al. 1989). Thermus thermophilus HB27 (ATCC BAA-163), on the other hand, was used as a thermophilic host for the expression of the protein at 60–70 ºC. The host, T. thermophilus was cultured from a 20% glycerol stock (maintained at -80 ºC) as described (Erasmus et al. 2014). Thermus broth [8 g L−1 tryptone (Biolab), 4 g L−1 yeast extract (Biolab), 3 g L−1 NaCl (Saarchem), and 2 M NaOH, pH 7.0] (Ramírez-Arcos et al. 1998), was used as the growth medium. The thermus broth medium was supplemented with 3 µg mL−1 bleomycin or 30 µg mL−1 kanamycin as required. Amplification of ABC transporter peptide-binding gene was carried out using the primers mentioned in Table 1. The PCR was performed in a total reaction volume of 50 µL, using a Thermal Cycler (PxE 0.2, Thermo Electron Corporation). The oligonucleotide primers (Primers 1 and 2) were designed to yield constructs that included the leader peptide and contained no His-Tag when expressed in pET-22b( +) and a N-terminal His-Tag when expressed with the pET-28b( +) vector.
Cloning and expression of protein
Plasmid pGEM®-T Easy (Promega) was used for sub-cloning of the gene into E. coli and plasmids pET-22b( +) and pET-28b( +) (Novagen) were used to express the gene in E. coli BL21(DE3). The pET-22b( +) vector was used for expression without any His-Tags by cloning into the vector between the EcoRI and NdeI sites and contains an ampicillin resistance. pET-28b( +) vector was used for expression with a N-terminal His-Tag by cloning into the vector between the EcoRI and NdeI sites that contains a kanamycin resistance gene. The recombinant vector map, designed by SnapGene software v5.0, was used for the expression of transgenes in pET-22b( +), pET-28b( +), pWUR and pMK184 as shown in Figs. 1 and 2. Plasmids pWUR (Brouns et al. 2005) and pMK184 (De Grado et al. 1999) were used to clone and express bleomycin resistance and kanamycin resistance gene, respectively by removing two of the three BamHI restriction sites in T. thermophilus as described (De Grado et al. 1999; Brouns et al. 2005) (Fig. 2). PCR amplification with oligonucleotide primers (primers 1, 2, 5, and 6) for cloning and expression are described in Table 1.
Purification and identification of ABC transporter, peptide-binding gene
The inserted plasmid was purified by excising the band at ± 1.9 kb on the agarose gel using a non-UV transilluminator (Dark Reader™ transilluminator DR-45 M, Clare Chemical Research) and extracted using the BioSpin Gel Extraction Kit (BioFlux), following the manufacturer’s instructions. The purified inserts were confirmed using Sanger sequencing (ABI Prism® Big Dye™ Terminator Cycle Sequencing Ready Reaction Kit V.3.1) as described by the manufacturer, and the primers used in the reaction are 3, 4, 7, and 8 (Table 1). The purified inserts were sequenced again using Sanger sequencing with primers 9, 10, 11, and 12 (Table 1) to confirm the nucleotide composition of the ABC transporter, peptide-binding gene.
Positive clones for the constructs in pET-22b( +) and pET-28b( +) as well as plasmids pWUR and pMK184 were identified through selection on an LB medium and Thermus broth, respectively, containing the required antibiotic. E. coli BL21 cells were incubated at 37 ºC and T. thermophilus HB27 at 70 ºC with aeration (200 rpm) until an OD600nm of approximately 0.8–1.0 was reached. Isopropyl β-D-1-Thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and cells were again incubated for an additional 4 h. Samples were collected every 2 h for the homogeneity of the ABC transporters, peptide-binding proteins present in each fraction using sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) analysis as described (Laemmli 1970).
Gold reduction and nanoparticle formation
The purified protein solutions were used in reducing gold (III) for nanoparticle production by adapting the procedure described (Erasmus et al. 2014). An aliquot of 100 µL of the purified protein was reacted with 100 µL gold (III) solution (HAuCl4; 0–10 mM), 100 µl methyl viologen (1 µM), 100 µL phosphate buffer (50 mM; pH 7.4), and 500 µL double distilled water. The reaction was activated by adding 100 µL sodium dithionite (46 µM) dissolved in sodium hydrogen carbonate (100 µM). The reaction was incubated and maintained for 24 h. The transition from a pink/purple to a blue colour indicated gold reduction and nanoparticle (Lazarides and Schatz 2000). A control sample was prepared by replacing the purified protein with 100 µL phosphate buffer (50 mM; pH 7.4). Denatured protein was tested for gold reduction and nanoparticle formation by boiling protein for 3 h, which replaced the purified active protein. A SpectraMax M2 UV–Vis Scanner (Labotec) was used to measure gold reduction and nanoparticle formation in the range of 300–800 nm. Gold reduction (HAuCl4) and nanoparticle formation were tested through the range of gold (III) concentrations (0, 0.5, 1.0, 2.0, 5.0, 7.5, and 10 mM) at 85 °C in the thermophilic host and 30 °C in the mesophilic host; the effect of temperature was tested at the optimum concentration of HAuCl4 (2 mM) through the incubation of 30, 37, 42, 55, 65, and 85 °C; and the effect of pH was also tested at the optimum concentration of HAuCl4 through pH levels of 3.6, 5.5, 7.5, and 9.0 at 85 °C and 30 °C in the thermophilic and mesophilic host, respectively.
Transmission electron microscopy and energy dispersive X-ray spectrometry
The size and morphology of the nanoparticles were assessed using transmission electron microscopy (TEM). A drop of the washed sample was placed on Formvar carbon-coated 200-mesh copper grids for analysis. After allowing the grid to air-dry, electron micrographs of the gold nanoparticles were taken with a JEOL TEM 1010, 60 kV TEM. Energy dispersive X-ray spectrometry (EDX) was used to identify the chemical composition of the particles using an EDAX DX4 energy dispersive X-ray spectrometry system, coupled to a 200 kV Philips CM 20 TEM.
Results and discussions
Purification and identification of ABC transporter, peptide-binding gene
Thermus scotoductus SA-01 is a well-known bacteria strain capable of reducing numerous compounds, including U (VI), Fe (III), Mn(IV), and Au (III) (Kieft et al. 1999). The inherent characteristic of the strain having several membrane proteins and cytoplasmic proteins which accept electrons through dissimilatory pathways gave T. scotoductus SA-01 the affinity to contribute to the gold reduction and nanoparticle production processes (Van Marwijk 2009; Gounder et al. 2011). Following N-terminal sequencing, an ABC transporter, peptide-binding protein from T. thermophilus (YP_144900) was identified (Van Marwijk 2009). However, the gene sequence, matches the ABC transporter, peptide-binding protein in the T. scotoductus SA-01 genome in the database, constructed by Gounder (2009) using pyrosequencing. The ABC transporter, peptide-binding protein has been identified as a reductase protein capable of reducing uranium (Cason et al. 2012), gold, chromate, and iron (Balkwill et al. 2004; Opperman and Van Heerden 2007; Gounder et al. 2011). Furthermore, the genome of T. scotoductus SA-01 has an average G + C content of approximately 69%, similar to that of T. thermophilus, and T. thermophilus genes are part of the T. scotoductus chromosome, but due to mutations that resulted in a loss of synteny between the chromosomes of the two strains, they became individual microbes (Gounder et al. 2011). Therefore, the peptide-binding protein in T. scotoductus gDNA was transformed into the thermophilic host T. thermophilus HB27 due to their similarity and the mesophilic host E. coli BL21 for protein expression in this study. The final elution profile of the purified protein showed distinct peaks in the clones pET-22b( +), pET-28b( +), pWUR, and pMK184 vectors for overexpression of the ABC transporter, peptide-binding protein. The overexpressed protein on the SDS-PAGE gel gave high levels of peaks at ± 70 kDa (Fig. 3A and B). There were no significant changes between the protein expressed in pET-22b( +), which had no His-Tags, and the protein expressed in pET-28b( +), which had an N-terminal His-Tag. However, the expression levels in the pWUR and pMK184 vectors were notably lower than the expression done with the pET vectors. This is probably due to the weaker PslpA promoter in T. thermophilus as compared to the strong T7 promoter system in E. coli.
A Expression of the ABC transporter, peptide-binding protein at ± 70 kDa. Lane 1: Precision Plus ProteinTM Standard; Lanes 2 and 6: negative controls of pET-22b( +) and pET-28b( +) respectively, in 4 h. Lanes 3–5: clone 7 in pET-22b( +) in 0, 2, and 4 h, respectively. Lanes 7–9: clone 9 in pET-28b( +), in 0, 2, and 4 h, respectively. B: Expression of the ABC transporter, peptide-binding protein of ± 70 kDa. Lane 1: Precision Plus ProteinTM Standard; Lanes 2 and 6: negative controls of pWUR and pMK184, respectively; Lanes 3–5: clone 10 (Construct 1), clone 2 (Construct 2), and clone 7 (Construct 3) in pWUR, respectively. Lanes 7 and 8: clone 4 (Construct 2 and 3) in pMK184, respectively
Gold reduction nanoparticle formation
Protein-mediated reduction and nanoparticle formation reactions were done using the purified protein and a UV–Vis spectrum was used to observe changes. The surface plasmon resonance obtained from the UV–Vis spectrum reveals that optimum nanoparticle formation occurs at concentrations ranging from 0.5 mM to 2 mM of HAuCl4 in proteins from both mesophilic [pET-22b( +) and pET-28b( +)] and thermophilic (pWUR and pMK184 vectors) hosts. Nanoparticle formations were notable in the absorbance around 520–560 nm in the protein reactions (Fig. 4A and B), similar to the UV–visible spectra wavelength (524 nm) reported by Monsalve et al. (2015). Nanoparticle formations from 5.0 to 10 mM were weak, as the curves slanted downward and nanoparticle formations may not be properly formed in the thermophilic host. On the other hand, there was no significant change in absorbance for the mesophilic host, indicating the particle form may not be nanosized, and there was no reaction at 0 mM, indicating no interference that could have influenced nanoparticle formation. The formation of nanostructures occurs when Au(III) is reduced to Au(0), thereby leading to the formation of nucleation sites that initiate the synthesis of nanoparticles (Tang and Hamley 2009).
UV–Vis absorption spectra of the surface plasmon resonance and protein reactions recorded for the overexpression of ABC transporter, peptide-binding protein at 30 °C and 85 °C in Escherichia coli BL21 and Thermus thermophilus HB27 hosts, respectively. (A and B): represent the different HAuCl4 concentrations, indicating gold nanoparticle formation at wavelengths 540–560 nm. (C and D): effect of different temperatures on gold nanoparticle formation with changes in the absorbance around 520–560 nm. (E and F): different pH values, gold nanoparticle formation changes are in the absorbance between 520–650 nm
The response of extracted proteins from mesophilic and thermophilic hosts to temperature differs in terms of gold reduction and nanoparticle production. Nanoparticle formation in mesophilic host proteins was observed to begin at 55 °C, with optimum formation at 65 °C and excessive chemical reduction at 85 °C as protein reaction gave a dark pink coloration at 65 °C, indicating polydispersed nanoparticles at absorbance ranging from 520 to 550 nm (Rajeshkumar et al. 2013) (Fig. 4C). Gold reduction and nanoparticle formation in thermophilic host are significantly pronounced at temperatures 65 ºC and 85 ºC with optimum formation at 85 °C, revealing through a change in the absorbance from 520 to 560 nm in the protein reactions (Fig. 4D), suggesting thermostable protein expression in the T. thermophilus HB27. The thermophilic host favours nanoparticle formation at higher temperatures, unlike the mesophilic host, whose biosynthesis efficiency decreases at high temperatures due to enzyme denaturation (El-Shanshoury et al. 2020). This corroborates the findings of Gericke and Pinches (2006), where an increase in temperature permits faster particle formation. Therefore, the biogenic synthesis of gold nanoparticles is temperature-dependent to attain an optimum ultrafine particulate and also depends on the host and environmental conditions.
The alteration of pH has significant effects on gold reduction and nanoparticle formations, as revealed through UV–Vis spectra (Fig. 4E and F). Nanoparticles were formed at all pH values, with optimal formation occurring at pH 5.5–9.0 for mesophilic host protein. This corresponds to the findings of Abdulazeem and Abd (2022), who reported the optimal pH for HAuCl4 salt reduction (10–3 mM) by E. coli at 37 °C to be 8.0. Thermophilic host proteins also formed nanoparticles at all pH values except 3.6, but the optimal nanoparticle formation occurred at pH 7.5 and 9.0, as indicated by the absorbance between 520 and 650 nm in the protein reactions. This suggests that protein constructs in T. thermophilus could perform optimally at higher pH levels, as the strain has been reported to grow optimally at a pH of 7.5 and at relatively high pH levels but not more than pH 9.6 (Oshima and Imahori 1974; Miyamoto et al. 2020). However, reports indicate that T. thermophilus's protein activity was high at pH 9.0 (Miyamoto et al. 2020). This supports the optimal nanoparticle formation in this study, which occurred at pH 7.5–9.0. For each parameter, the effect of denatured proteins on gold reduction and nanoparticle formation was examined. Denatured proteins do not effectively initiate gold reduction and nanoparticle formation under all conditions, as there were no significant changes in absorbance (520–560 nm) in the protein reactions and corresponding blank reactions (results not presented). This is not surprising because proteins tend to lose activity when subjected to high temperatures as a result of denaturation.
TEM micrographs and EDX measurements of the nanoparticle formation
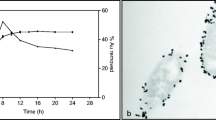
Transmission electron microscopic and energy-dispersive X-ray spectroscopic analyses were carried out. These analyses elucidate the formation and spatial distribution of nanoparticles. This is associated with the purified ABC transporter peptide-binding protein constructs from cloned E. coli and T. thermophilus. The TEM micrographs and EDX confirmed that the particles formed were in the nanosize range. Nanosized particles were formed with spherical gold shapes in the range of 0.5 mM to 2.0 mM concentrations of gold in both hosts (Fig. 5). However, no significant biosynthesized formation was observed at the remaining concentrations as samples showed a larger mean particle size; hence, the micrographs were not included in this study. The micro-diffraction pattern for gold is noticeable in Fig. 5C and D, with several shapes and sizes formed, mostly spherical, at 1.0 mM gold concentration, but as the concentration increases, particle formation increases in size which may be larger than nanosize range. However, no observable differences were obtained using similar gold concentrations with the protein expressed in a thermophilic host, confirming that biogenic nanoparticle formation using microbes is concentration dependent. On the other hand, abiotic control experiments without purified protein did not demonstrate the presence of HAuCl4 nanoparticle formation in the solution either by a colour change which is an indicator or by TEM micrograph (Daruich De Souza et al. 2019). Similarly, TEM micrographs confirmed no interference for the assays with a 0 mM HAuCl4 solution.
The transmission electron micrographs confirmed that particles were indeed formed at each temperature, with improved formation at the optimal temperatures of the mesophilic (30 ºC) and thermophilic (85 ºC) hosts, and that all the particles are in the nano-size range and shapes but are not homogeneous (Fig. 6). At 30 ºC, 37 ºC, and 42 ºC, proteins from the mesophilic host exhibit well-defined spherical-shaped particles. However, as the temperature rises, the size of the particles increases, and at 85 ºC, the shape becomes less defined. The TEM micrographs confirmed significant biosynthesized nanoparticle formation at 65 ºC and 85 ºC (Fig. 6C and D) in the thermophilic host protein which corresponded to the UV–Vis absorption spectra. The formation of nanoparticles at high temperatures indicates that the ABC transporter peptide-binding gene encoding thermostability and resistance to antibiotics was not distorted, and the proteins are stable at high temperatures. It has been confirmed that enzymes and proteins are stable in thermophilic microorganisms at high temperatures and under other adverse conditions, including extreme pH, concentrated detergents, and chaotropic agents (Politi et al. 2016). This could be advantageous in the formation of nanoparticles using thermophilic microorganisms.
The pH range of 3.6–9.0 used to test the impact of pH on gold reduction and nanoparticle formation showed that pH can affect the microbial biosynthesis of gold nanoparticles (Fig. 7), which is also confirmed in the study of He et al. (2007). In the mesophilic host proteins, nanoparticles were formed at all pH values, with optimum formation at pH 5.5–9.0 and the response of proteins is observed at absorbance 540–550 nm (Fig. 7A, B, and C1). However, the effect of pH on gold reduction and nanoparticle formation was significant at pH 7.5 and 9.0 (Fig. 7D and E1) with corresponding colour change and changes in the UV–Vis spectra absorbance 520 and 650 nm in thermophilic host proteins. Different pH values correlate with the proton concentration of a solution, thereby influencing nanoparticle morphology. Incubation at higher pH values must have led to the formation of more spherical nanoparticles (Erasmus et al. 2014). Politi et al. (2016) also observed optimal activities of arsenate reductase from T. thermophilus at pH 8.0 and 9.0, indicating that these pH values could optimize the formation of T. thermophilus biosynthesized nanoparticles. The morphology and chemical composition of nanoparticles is influenced by biosynthetic conditions, including pH, temperature, and HAuCl4 concentration (Sun and Xia 2002). As a result, the nanoparticles formed at the range of temperatures (30–85 ºC), pH (3.6–9.0) and concentrations of HAuCl4 solution using the ABC transporter, peptide-binding protein constructs in E. coli and T. thermophilus were further examined by EDX analysis, which confirmed the nanoparticles to be gold as shown in Fig. 8. However, there are limitations in synthesising monodisperse nanoparticles using the protein constructs from E. coli and T. thermophilus. This is not peculiar to our study. It has been reported in other studies that synthesising monodisperse nanoparticles with long-term stability and exceptional mass activity is challenging due to the uneven reduction rates of metal precursors (Lao et al. 2021; Kar et al. 2024). However, the use of the one-step oil bath method has been proposed to improve the metals reduction process and reaction intermediates (Lao et al. 2021). EDX spectrum diffraction can characterize the chemical or elemental components of nanoparticles (Menon et al. 2017), thus, it was used in this study to determine whether the gold nanoparticles produced were gold. Carbon and copper were also detected in the samples due to the TEM grids, as the analysis was done on Formvar carbon-coated 200-mesh copper grids.
TEM micrographs for the pH values; A, B, and C1 with the corresponding selected-area electron micro-diffraction pattern for pH 9.0 (C2) for Escherichia coli BL21 host protein at the optimum temperature (30 °C). D and E1 are pH values of 7.5 and 9.0 respectively for Thermus thermophilus HB27 host protein at 85 ºC, with the corresponding selected-area electron micro-diffraction pattern (E2). Square-shaped panels represent the magnification
Conclusion
The ABC transporter, peptide-binding protein, from T. scotoductus SA-01 was cloned and expressed in a mesophilic E. coli and thermophilic T. thermophilus host. The purified ABC transporter, peptide-binding protein, successfully reduced gold (III) to gold (0) nanoparticles, which were validated using various abiotic parameters. However, the formation of gold nanoparticles using protein constructs from E. coli and T. thermophilus was not monodispersed, suggesting further research might be necessary to optimise the formation. Monodisperse nanoparticles hold great promise in several applications, including biological, medical, and engineering applications, due to their unique natural compatibility characteristics. Overall, the optimum nanoparticles were observed at higher pH values (5.5 to 9.0), lower gold (III) concentrations (0.5 to 2.0 mM), and at room temperature but not more than 45 ºC in the mesophilic host but higher temperature (85 ºC) in the thermophilic host, indicating the significance of temperature in both hosts for expression, folding, and activity of the ABC transporter protein. Furthermore, transmission electron microscopy and energy-dispersive X-ray spectrometry confirmed that the nanoparticles were associated with the purified protein constructs from the cloned hosts. Findings from this study suggest the level of effectiveness of the purified ABC transporter peptide binding protein in the reduction of gold and formation of nanoparticles, especially when expressed in thermophilic T. thermophilus. The technique employed in this study could further be optimized for commercial or large-scale synthesis of nanoparticles at high temperatures using thermophilic organisms for various biological applications of nanoparticles, including drug delivery, diagnosis, catalysis, sensors, electronics, and bioremediation of gold-bearing waste.
Data availability
All data generated or analysed during this study is included in this article.
Abbreviations
- Ag:
-
Silver
- Au:
-
Gold (Au)
- DNA:
-
Deoxyribonucleic acid
- EDX:
-
Energy dispersive X-ray spectrometry
- IPTG:
-
Isopropyl β-D-1-Thiogalactopyranoside
- IPTG:
-
Isopropyl β-D-1-Thiogalactopyranoside
- LB:
-
Luria-Bertani
- NBDs:
-
Nucleotide-binding domains
- OD:
-
Optical density
- PCR:
-
Polymerase chain reaction
- PH:
-
Potential of hydrogen
- Pt:
-
Platinum
- RPM:
-
Revolutions Per Minute
- SDS-PAGE:
-
Sodium dodecyl-sulfate polyacrylamide gel electrophoresis
- TEM:
-
Transmission electron microscopy
- TMDs:
-
Two transmembrane domains
- TYG:
-
Tryptone, Yeast Extract, Glucose
- UV:
-
Ultraviolet
References
Abdulazeem L, Abd FG (2022) Optimization of gold nanoparticles synthesis from local isolate of E. coli and their activity as antibacterial. AIP Conf Proc 2398:040003. https://doi.org/10.1063/5.0095231
Asnag GM, Oraby AH, Abdelghany AM (2019) Green synthesis of gold nanoparticles and its effect on the optical, thermal and electrical properties of carboxymethyl cellulose. Compos B Eng 172:436–446. https://doi.org/10.1016/j.compositesb.2019.05.044
Balkwill DL, Kieft TL, Tsukuda T, Kostandarithes HM, Onstott TC, Macnaughton S, Bownas J, Fredrickson JK (2004) Identification of iron-reducing Thermus strains as Thermus scotoductus. Extremophiles 8:37–44. https://doi.org/10.1007/s00792-003-0357-0
Bhatwa A, Wang W, Hassan YI, Abraham N, Li XZ, Zhou T (2021) Challenges associated with the formation of recombinant protein inclusion bodies in Escherichia coli and strategies to address them for industrial applications. Front Bioeng Biotechnol 9:630551. https://doi.org/10.3389/fbioe.2021.630551
Brouns SJJ, Wu H, Akerboom J, Turnbull AP, De Vos WM, Van Der Oost J (2005) Engineering a selectable marker for hyperthermophiles. J Biol Chem 280:11422–11431. https://doi.org/10.1074/jbc.M413623200
Cai J, Gros P (2003) Overexpression, purification, and functional characterization of ATP-binding cassette transporters in the yeast, Pichia pastoris. Biochem Biophys Acta (BBA) - Biomembranes 1610:63–76. https://doi.org/10.1016/S0005-2736(02)00718-6
Campbell FW, Compton RG (2010) The use of nanoparticles in electroanalysis: An updated review. Anal Bioanal Chem 396:241–259. https://doi.org/10.1007/s00216-009-3063-7
Carr JF, Danziger ME, Huang AL, Dahlberg AE, Gregory ST (2015) Engineering the genome of Thermus thermophilus using a Counter selectable Marker. J Bacteriol 197:1135. https://doi.org/10.1128/jb.02384-14
Cason ED, Piater LA, van Heerden E (2012) Reduction of U(VI) by the deep subsurface bacterium, Thermus scotoductus SA-01, and the involvement of the ABC transporter protein. Chemosphere 86:572–577. https://doi.org/10.1016/j.chemosphere.2011.10.006
De Grado M, Castán P, Berenguer J (1999) A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42:241–245. https://doi.org/10.1006/plas.1999.1427
De Souza CD, Nogueira BR, Rostelato MECM (2019) Review of the methodologies used in the synthesis gold nanoparticles by chemical reduction. J Alloys Compd 798:714–740. https://doi.org/10.1016/j.jallcom.2019.05.153
Dean M, Hamon Y, Chimini G (2001) The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res 42:1007–1017. https://doi.org/10.1016/S0022-2275(20)31588-1
El-Shanshoury A, Ebeid E, Elsilk S, Mohamed S, Ebeid M (2020) Biogenic synthesis of gold nanoparticles by bacteria and utilization of the chemical fabricated for diagnostic performance of viral hepatitis C Virus-NS4. Lett Appl NanoBioSci 9:1395–1408. https://doi.org/10.33263/LIANBS93.13951408
Erasmus M, Cason ED, Van Marwijk J, Botes E, Gericke M, Van Heerden E (2014) Gold nanoparticle synthesis using the thermophilic bacterium Thermus scotoductus SA-01 and the purification and characterization of its unusual gold reducing protein. Gold Bull 47:245–253. https://doi.org/10.1007/s13404-014-0147-8
Gounder K (2009) Genome sequencing of the extremophile Thermus scotoductus SA-01 and expression of selected genes. University of Free State, Bloemfontein
Gounder K, Brzuszkiewicz E, Liesegang H, Wollherr A, Daniel R, Gottschalk G, Reva O, Kumwenda B, Srivastava M, Bricio C, Berenguer J, van Heerden E, Litthauer D (2011) Sequence of the hyperplastic genome of the naturally competent Thermus scotoductus SA-01. BMC Genomics 12:577. https://doi.org/10.1186/1471-2164-12-577
He S, Guo Z, Zhang Y, Zhang S, Wang J, Gu N (2007) Biosynthesis of gold nanoparticles using the bacteria Rhodopseudomonas capsulata. Mater Lett 61:3984–3987. https://doi.org/10.1016/j.matlet.2007.01.018
Henne A, Brüggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, Jacobi C, Starkuviene V, Schlenczeck S, Dencker S, Huber R, Klenk HP, Kramer W, Merkl R, Gottschalk G, Fritz HJ (2004) The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol 22:547–553. https://doi.org/10.1038/nbt956
Kar N, McCoy M, Wolfe J, Bueno SL, Shafei IH, Skrabalak SE (2024) Retrosynthetic design of core–shell nanoparticles for thermal conversion to monodisperse high-entropy alloy nanoparticles. Nat Synthesis 3:175–184. https://doi.org/10.1038/s44160-023-00409-0
Khan I, Saeed K, Khan I (2019) Nanoparticles: Properties, applications and toxicities. Arabian J Chem 12:908–931. https://doi.org/10.1016/j.arabjc.2017.05.011
Kieft TL, Fredrickson JK, Onstott TC, Gorby YA, Kostandarithes HM, Bailey TJ, Kennedy DW, Li SW, Plymale AE, Spadoni CM, Gray MS (1999) Dissimilatory reduction of Fe(III) and other electron acceptors by a Thermus isolate. Appl Environ Microbiol 65:1214–1221. https://doi.org/10.1128/AEM.65.3.1214-1221.1999
Ko S, Huh C (2019) Use of nanoparticles for oil production applications. J Pet Sci Eng 172:97–114. https://doi.org/10.1016/j.petrol.2018.09.051
Korbekandi H, Iravani S, Abbasi S (2009) Production of nanoparticles using organisms. Crit Rev Biotechnol 29:279–306. https://doi.org/10.3109/07388550903062462
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Lao X, Yang M, Sheng X, Sun J, Wang Y, Zheng D, Pang M, Fu A, Li H, Guo P (2021) Monodisperse PdBi nanoparticles with a face-centered cubic structure for highly efficient ethanol oxidation. ACS Appl Energy Mat 5:1282–1290. https://doi.org/10.1021/acsaem.1c03703
Lasa I, Castón JR, Fernández-Herrero LA, de Pedro MA, Berenguer J (1992) Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol Microbiol 6:1555–1564. https://doi.org/10.1111/j.1365-2958.1992.tb00877.x
Lazarides AA, Schatz GC (2000) DNA-linked metal nanosphere materials: structural basis for the optical properties. J Phys Chem B 104:460–467. https://doi.org/10.1021/jp992179+
Lee JE, Lee N, Kim T, Kim J, Hyeon T (2011) Multifunctional mesoporous silica nanocomposite nanoparticles for theranostic applications. Acc Chem Res 44:893–902. https://doi.org/10.1021/ar2000259
Li X, Xu H, Chen ZS, Chen G (2011) Biosynthesis of nanoparticles by microorganisms and their applications. J Nanomater 2011:270974. https://doi.org/10.1155/2011/270974
Menon S, Rajeshkumar S, Kumar VS (2017) A review on biogenic synthesis of gold nanoparticles, characterization, and its applications. Res-Efficient Techno 3:516–527. https://doi.org/10.1016/j.reffit.2017.08.002
Miyamoto T, Moriya T, Homma H, Oshima T (2020) Enzymatic properties and physiological function of glutamate racemase from Thermus thermophilus. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1868:140461. https://doi.org/10.1016/j.bbapap.2020.140461
Monsalve K, Roger M, Gutierrez-Sanchez C, Ilbert M, Nitsche S, Byrne-Kodjabachian D, Marchi V, Lojou E (2015) Hydrogen bioelectrooxidation on gold nanoparticle-based electrodes modified by Aquifex aeolicus hydrogenase: Application to hydrogen/oxygen enzymatic biofuel cells. Bioelectrochemistry 106:47–55. https://doi.org/10.1016/j.bioelechem.2015.04.010
Opperman DJ, Van Heerden E (2007) Aerobic Cr(VI) reduction by Thermus scotoductus strain SA-01. J Appl Microbiol 103:1907–1913. https://doi.org/10.1111/j.1365-2672.2007.03429.x
Opperman DJ, Piater LA, Van Heerden E (2008) A novel chromate reductase from Thermus scotoductus SA-01 related to old yellow enzyme. J Bacteriol 190:3076–3082. https://doi.org/10.1128/jb.01766-07
Oshima T, Imahori K (1974) Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol 24:102–112. https://doi.org/10.1099/00207713-24-1-102
Politi J, Spadavecchia J, Fiorentino G, Antonucci I, De Stefano L (2016) Arsenate reductase from Thermus thermophilus conjugated to polyethylene glycol-stabilized gold nanospheres allow trace sensing and speciation of arsenic ions. J R Soc Interface 13:20160629. https://doi.org/10.1098/rsif.2016.0629
Popovic M, Zaja R, Loncar J, Smital T (2010) A novel ABC transporter: The first insight into zebrafish (Danio rerio) ABCH1. Mar Environ Res 69:S11–S13. https://doi.org/10.1016/j.marenvres.2009.10.016
Rajeshkumar S, Malarkodi C, Gnanajobitha G, Paulkumar K, Vanaja M, Kannan C, Annadurai G (2013) Seaweed-mediated synthesis of gold nanoparticles using Turbinaria conoides and its characterization. J Nanostructure Chem 3:1–7. https://doi.org/10.1186/2193-8865-3-44
Ramírez-Arcos S, Fernández-Herrero LA, Marín I, Berenguer J (1998) Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J Bacteriol 180:3137–3143. https://doi.org/10.1128/jb.180.12.3137-3143.1998
Rea PA (2007) Plant ATP-binding cassette transporters.https://doi.org/10.1146/annurev.arplant.57.032905.105406.58:347-375
Salata OV (2004) Applications of nanoparticles in biology and medicine. J Nanobiotechnol 2:3. https://doi.org/10.1186/1477-3155-2-3
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
Shamaila S, Sajjad AKL, Ryma NA, Farooqi SA, Jabeen N, Majeed S, Farooq I (2016) Advancements in nanoparticle fabrication by hazard free eco-friendly green routes. Appl Mater Today 5:150–199. https://doi.org/10.1016/j.apmt.2016.09.009
Shnoudeh AJ, Hamad I, Abdo RW, Qadumii L, Jaber AY, Surchi HS, Alkelany SZ (2019) Synthesis, characterization, and applications of metal nanoparticles. Biomater Bionanotechnol 527–612. https://doi.org/10.1016/B978-0-12-814427-5.00015-9
Singh P, Kim YJ, Zhang D, Yang DC (2016) Biological synthesis of nanoparticles from plants and microorganisms. Trends Biotechnol 34:588–599. https://doi.org/10.1016/j.tibtech.2016.02.006
Sinha S, Pan I, Chanda P, Sen SK (2009) Nanoparticles fabrication using ambient biological resources. J Appl Biosci 19:1113–1130
Sun Y, Xia Y (2002) Shape-controlled synthesis of gold and silver nanoparticles. Science 298:2176–2179. https://doi.org/10.1126/science.1077229
Tang T, Hamley IW (2009) Multiple morphologies of gold nano-plates by high-temperature polyol syntheses. Colloid Surf a: Physichochem Eng Asp 336:1–7. https://doi.org/10.1016/j.colsurfa.2008.11.008
Turner P, Mamo G, Karlsson EN (2007) Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb Cell Fact 6:9. https://doi.org/10.1186/1475-2859-6-9
Ullah H, Khan I, Yamani ZH, Qurashi A (2017) Sonochemical-driven ultrafast facile synthesis of SnO2 nanoparticles: growth mechanism structural electrical and hydrogen gas sensing properties. Ultrason Sonochem 34:484–490. https://doi.org/10.1016/j.ultsonch.2016.06.025
Van Marwijk J (2009) Biological synthesis of gold nanoparticles by Thermus scotoductus SA-01. Free State University, Bloemfontein
Yezhelyev MV, Gao X, Xing Y, Al-Hajj A, Nie S, O’Regan RM (2006) Emerging use of nanoparticles in diagnosis and treatment of breast cancer. Lancet Oncol 7:657–667. https://doi.org/10.1016/S1470-2045(06)70793-8
Acknowledgements
The authors acknowledge the support of the National Research Foundation, the Oppenheimer Memorial Trust, the Ernst and Ethel Eriksen Trust, and the Technology Innovation Agency (TIA), South Africa, in this study.
Funding
Open access funding provided by University of the Free State. Whitehead Scientific, BioPAD (TIA), and National Research Foundation (NRF) research grants all contributed to the financial support for this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interest
The authors declare there is no conflict of interest associated with this publication. All authors have read and agreed to the publication of the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Erasmus, M., Idris, O.A., Adetunji, A.I. et al. Biogenic synthesis and characterization of gold nanoparticles using transformed mesophilic Escherichia coli BL21 and thermophilic Thermus thermophilus HB27. Biologia (2024). https://doi.org/10.1007/s11756-024-01733-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11756-024-01733-2